4TQ3
 
 | | Structure of a UbiA homolog from Archaeoglobus fulgidus bound to GPP and Mg2+ | | Descriptor: | GERANYL DIPHOSPHATE, MAGNESIUM ION, Prenyltransferase | | Authors: | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4076 Å) | | Cite: | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
4TT6
 
 | | Crystal structure of ATAD2A bromodomain double mutant N1063A-Y1064A in apo form | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TTE
 
 | | Crystal structure of ATAD2A bromodomain complexed with methyl 3-amino-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzoate | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-20 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TNA
 
 | |
4U2R
 
 | | Crystal structure of the GLUR2 ligand binding core (S1S2J, flip variant) in the apo state | | Descriptor: | Glutamate receptor 2, SULFATE ION | | Authors: | Duerr, K.L, Chen, L, Gouaux, E. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4114 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U01
 
 | | HCV NS3/4A serine protease in complex with 6570 | | Descriptor: | (2S,3aS,10Z,11aS,12aR)-2-({8-fluoro-7-methoxy-2-[4-(propan-2-yl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)-5-methyl-N-[(1-methylcyclopropyl)sulfonyl]-4,14-dioxo-1,2,3,3a,4,5,6,7,8,9,11a,12,13,14-tetradecahydro-12aH-cyclopropa[m]pyrrolo[1,2-c][1,3,6]triazacyclotetradecine-12a-carboxamide, CHLORIDE ION, NS4A protein, ... | | Authors: | Parsy, C.C, Alexandre, F.-R, Brandt, G, Caillet, C, Chaves, D, Derock, M, Gloux, D, Griffon, Y, Lallos, L.B, Leroy, F, Liuzzi, M, Loi, A.-G, Mayes, B, Moulat, L, Moussa, A, Chiara, M, Roques, V, Rosinovsky, E, Seifer, M, Stewart, A, Wang, J, Standring, D, Surleraux, D. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and structural diversity of the hepatitis C virus NS3/4A serine protease inhibitor series leading to clinical candidate IDX320.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4U0O
 
 | | Crystal structure of Thermosynechococcus elongatus Lipoyl Synthase 2 complexed with MTA and DTT | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 5'-DEOXY-5'-METHYLTHIOADENOSINE, IRON/SULFUR CLUSTER, ... | | Authors: | Harmer, J.E, Hiscox, M.J, Dinis, P.C, Sandy, J, Roach, P.L. | | Deposit date: | 2014-07-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions.
Biochem.J., 464, 2014
|
|
4U5C
 
 | | Crystal structure of GluA2, con-ikot-ikot snail toxin, partial agonist FW and postitive modulator (R,R)-2b complex | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Con-ikot-ikot, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.6883 Å) | | Cite: | X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism.
Science, 345, 2014
|
|
4U6D
 
 | | Zg3615, a family 117 glycoside hydrolase in complex with beta-3,6-anhydro-L-galactose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-beta-L-galactopyranose, CALCIUM ION, ... | | Authors: | Ficko-Blean, E. | | Deposit date: | 2014-07-28 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural investigation of two paralogous glycoside hydrolases from Zobellia galactanivorans: novel insights into the evolution, dimerization plasticity and catalytic mechanism of the GH117 family.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UAI
 
 | | Crystal structure of CXCL12 in complex with inhibitor | | Descriptor: | 1-phenyl-3-[4-(1H-tetrazol-5-yl)phenyl]urea, SULFATE ION, Stromal cell-derived factor 1 | | Authors: | Smith, E.W, Chen, Y. | | Deposit date: | 2014-08-09 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of a Novel Small Molecule Ligand Bound to the CXCL12 Chemokine.
J.Med.Chem., 57, 2014
|
|
4U6B
 
 | | Zg3597, a family 117 glycoside hydrolase, produced by the marine bacterium Zobellia galactanivorans | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Ficko-Blean, E. | | Deposit date: | 2014-07-28 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural investigation of two paralogous glycoside hydrolases from Zobellia galactanivorans: novel insights into the evolution, dimerization plasticity and catalytic mechanism of the GH117 family.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4U7K
 
 | | Crystal structure of the Clostridium histolyticum colH collagenase polycystic kidney disease-like domain 2a in the presence of calcium at 1.9 Angstrom resolution | | Descriptor: | CALCIUM ION, ColH protein | | Authors: | Bauer, R, Janowska, K, Sakon, J, Matsushita, O, Latimer, E. | | Deposit date: | 2014-07-31 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structures of three polycystic kidney disease-like domains from Clostridium histolyticum collagenases ColG and ColH.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UOG
 
 | | Crystallographic structure of nucleoside diphosphate kinase from Litopenaeus vannamei complexed with dCDP | | Descriptor: | DEOXYCYTIDINE DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Lopez-Zavala, A.A, Stojanoff, V, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2014-06-03 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Nucleoside Diphosphate Kinase from Pacific Shrimp (Litopenaeus Vannamei) in Binary Complexes with Purine and Pyrimidine Nucleoside Diphosphates
Acta Crystallogr.,Sect.F, 79, 2014
|
|
4UOF
 
 | | Crystallographic structure of nucleoside diphosphate kinase from Litopenaeus vannamei complexed with dADP | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Lopez-Zavala, A.A, Stojanoff, V, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2014-06-03 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structure of Nucleoside Diphosphate Kinase from the Pacific Shrimp (Litopenaeus Vannamei) in Binary Complexes with Purine and Pyrimidine Nucleoside Diphosphates
Acta Crystallogr.,Sect.F, 79, 2014
|
|
4UN0
 
 | | Crystal structure of the human CDK12-cyclinK complex | | Descriptor: | CYCLIN-DEPENDENT KINASE 12, CYCLIN-K | | Authors: | Dixon Clarke, S.E, Elkins, J.M, Pike, A.C.W, Chaikuad, A, Goubin, S, Krojer, T, Sorrell, F.J, Nowak, R, Williams, E, Kopec, J, Mahajan, R.P, Burgess-Brown, N, Carpenter, E.P, Knapp, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-05-22 | | Release date: | 2014-06-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of the Cdk12/Cyck Complex with AMP-Pnp Reveal a Flexible C-Terminal Kinase Extension Important for ATP Binding.
Sci.Rep., 5, 2015
|
|
4V3D
 
 | | The CIDRa domain from HB3var03 PfEMP1 bound to endothelial protein C receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOTHELIAL PROTEIN C RECEPTOR, ... | | Authors: | Lau, C.K.Y, Turner, L, Jespersen, J.S, Lowe, E.D, Petersen, B, Wang, C.W, Petersen, J.E.V, Lusingu, J, Theander, T.G, Lavstsen, T, Higgins, M.K. | | Deposit date: | 2014-10-17 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Conservation Despite Huge Sequence Diversity Allows Epcr Binding by the Pfemp1 Family Implicated in Severe Childhood Malaria.
Cell Host Microbe., 17, 2015
|
|
4U0P
 
 | | The Crystal Structure of Lipoyl Synthase in Complex with S-Adenosyl Homocysteine | | Descriptor: | IRON/SULFUR CLUSTER, Lipoyl synthase 2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Harmer, J.E, Hiscox, M.J, Sandy, J, Dinis, P.C, Roach, P.L. | | Deposit date: | 2014-07-13 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions.
Biochem.J., 464, 2014
|
|
9CGT
 
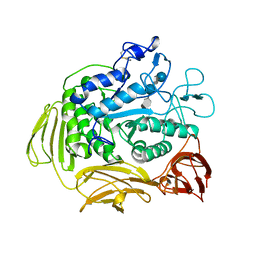 | | STRUCTURE OF CYCLODEXTRIN GLYCOSYLTRANSFERASE COMPLEXED WITH A THIO-MALTOPENTAOSE | | Descriptor: | CALCIUM ION, PROTEIN (CYCLODEXTRIN-GLYCOSYLTRANSFERASE), alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4-thio-alpha-D-glucopyranose-(1-4)-1-thio-alpha-D-glucopyranose | | Authors: | Schmidt, A.K, Schulz, G.E. | | Deposit date: | 1998-09-27 | | Release date: | 1998-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate binding to a cyclodextrin glycosyltransferase and mutations increasing the gamma-cyclodextrin production.
Eur.J.Biochem., 255, 1998
|
|
8V4M
 
 | | CCP5 in complex with microtubules class3 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-29 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 2024
|
|
4TSZ
 
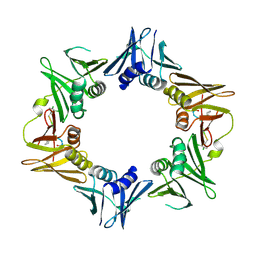 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa with ligand | | Descriptor: | ACE-GLN-ALC-ASP-LEU-ZCL peptide, DNA polymerase III subunit beta | | Authors: | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | Deposit date: | 2014-06-19 | | Release date: | 2014-09-10 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
8V4K
 
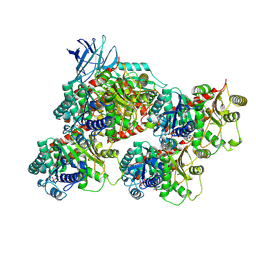 | | CCP5 in complex with microtubules class1 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-29 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 2024
|
|
9G13
 
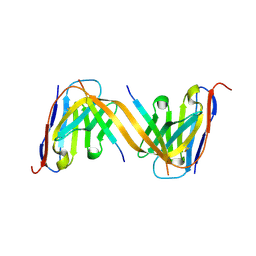 | | VHH H3-2 in complex with Tau C-terminal peptide | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau, VHH H3-2 | | Authors: | Dupre, E, Landrieu, I, Danis, C, Hanoulle, X, Mortelecque, J. | | Deposit date: | 2024-07-09 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | VHH H3-2 in complex with Tau C-terminal peptide
To Be Published
|
|
4ZIF
 
 | | Crystal Structure of core/latch dimer of Bax in complex with BimBH3mini | | Descriptor: | Apoptosis regulator BAX, Bcl-2-like protein 11 | | Authors: | Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
8VIW
 
 | | Cryo-EM structure of heparosan synthase 2 from Pasteurella multocida with polysaccharide in the GlcNAc-T active site | | Descriptor: | Heparosan synthase B, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Krahn, J.M, Pedersen, L.C, Liu, J, Stancanelli, E, Borgnia, M, Vivarette, E. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and Functional Analysis of Heparosan Synthase 2 from Pasteurella multocida to Improve the Synthesis of Heparin
Acs Catalysis, 14, 2024
|
|
8V4L
 
 | | CCP5 in complex with microtubules class2 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-29 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 2024
|
|
