6TN5
 
 | | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures | | Descriptor: | CHLORIDE ION, Heat shock protein HSP 90-alpha, ~{N}-(4-aminocarbonylphenyl)-~{N}-methyl-2,4-bis(oxidanyl)benzamide | | Authors: | Baker, L.M, Aimon, A, Murray, J.B, Surgenor, A.E, Matassova, N, Roughley, S.D, von Delft, F, Hubbard, R.E. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.165 Å) | | Cite: | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures
Commun Chem, 2020
|
|
2GF2
 
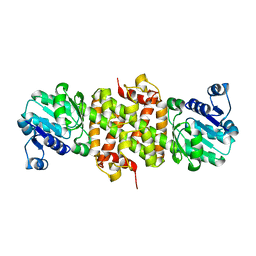 | | Crystal structure of human hydroxyisobutyrate dehydrogenase | | Descriptor: | 3-hydroxyisobutyrate dehydrogenase | | Authors: | Papagrigoriou, E, Salah, E, Turnbull, A.P, Smee, C, Burgess, N, Gileadi, O, von Delft, F, Gorrec, F, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Edwards, A.M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-21 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of human hydroxyisobutyrate dehydrogenase
To be Published
|
|
6FVT
 
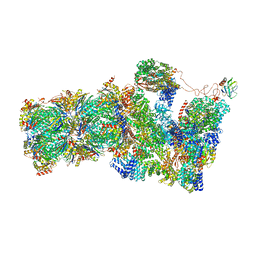 | | 26S proteasome, s1 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
8R04
 
 | | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active Site Inhibitor Cystargolide A | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6QZB
 
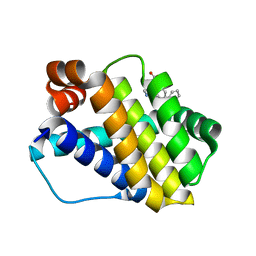 | | Structure of Mcl-1 in complex with compound 8d | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(2-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
8D43
 
 | |
1KH4
 
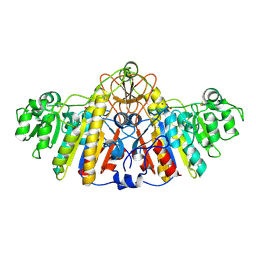 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D330N) IN COMPLEX WITH PHOSPHATE | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
2WID
 
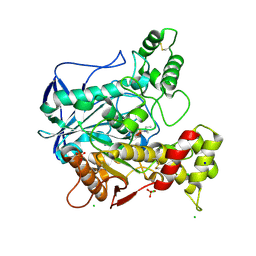 | | NONAGED FORM OF HUMAN BUTYRYLCHOLINESTERASE INHIBITED BY TABUN ANALOGUE TA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Carletti, E, Aurbek, N, Gillon, E, Loiodice, M, Nicolet, Y, Fontecilla, J, Masson, P, Thiermann, H, Nachon, F, Worek, F. | | Deposit date: | 2009-05-11 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-activity analysis of aging and reactivation of human butyrylcholinesterase inhibited by analogues of tabun.
Biochem. J., 421, 2009
|
|
6QW1
 
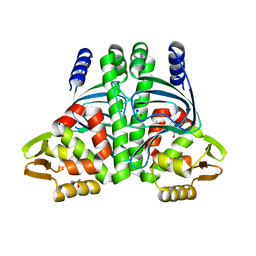 | | The Transcriptional Regulator PrfA-L140F mutant from Listeria Monocytogenes | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, SODIUM ION | | Authors: | Hall, M, Grundstrom, C, Hansen, S, Brannstrom, K, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes.
J.Bacteriol., 202, 2020
|
|
8OUC
 
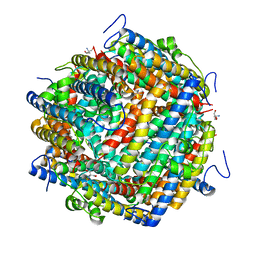 | | Escherichia coli DPS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA protection during starvation protein | | Authors: | Kovalenko, V.V, Loiko, N.G, Tereshkina, K.B, Tereshkin, E.V, Chulichkov, A.L, Popov, A.N, Krupyanskii, Y.F. | | Deposit date: | 2023-04-22 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Escherichia coli DPS
To be published
|
|
6D1Y
 
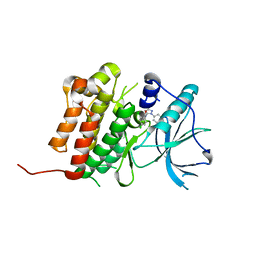 | | Crystal structure of Tyrosine-protein kinase receptor in complex with 2,4-dichloro-N-(3-methyl-1-phenyl-1H-pyrazol-5-yl)benzamide Inhibitor | | Descriptor: | 2,4-dichloro-N-(3-methyl-1-phenyl-1H-pyrazol-5-yl)benzamide, High affinity nerve growth factor receptor | | Authors: | Greasley, S.E, Johnson, E, Kraus, M.L, Cronin, C.N. | | Deposit date: | 2018-04-12 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of Allosteric, Potent, Subtype Selective, and Peripherally Restricted TrkA Kinase Inhibitors.
J. Med. Chem., 62, 2019
|
|
8P07
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with 5-((3-(4H-1,2,4-triazol-4-yl)phenyl)amino)-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidine-3-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, 7-(cyclopropylamino)-5-[[3-(1,2,4-triazol-4-yl)phenyl]amino]pyrazolo[1,5-a]pyrimidine-3-carbonitrile, Casein kinase II subunit alpha, ... | | Authors: | Kraemer, A, Ong, H.W, Yang, X, Bown, J.W, Chang, E, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | More than an Amide Bioisostere: Discovery of 1,2,4-Triazole-containing Pyrazolo[1,5- a ]pyrimidine Host CSNK2 Inhibitors for Combatting beta-Coronavirus Replication.
J.Med.Chem., 67, 2024
|
|
8I9R
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State 5S RNP | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, 60S ribosomal protein L16-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
2WMJ
 
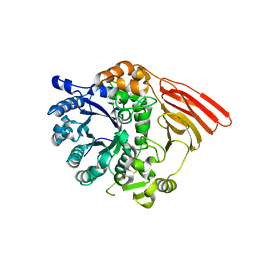 | | Crystal structure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 (Sp3GH98) in complex with the B-trisaccharide blood group antigen. | | Descriptor: | FUCOLECTIN-RELATED PROTEIN, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Higgins, M.A, Whitworth, G.E, El Warry, N, Randriantsoa, M, Samain, E, Burke, R.D, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differential Recognition and Hydrolysis of Host Carbohydrate-Antigens by Streptococcus Pneumoniae Family 98 Glycoside Hydrolases.
J.Biol.Chem., 284, 2009
|
|
6DZW
 
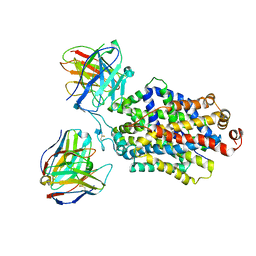 | | Cryo-EM structure of the ts2-inactive human serotonin transporter in complex with paroxetine and 15B8 Fab and 8B6 ScFv | | Descriptor: | 15B8 antibody heavy chain, 15B8 antibody light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coleman, J.A, Yang, D, Gouaux, E. | | Deposit date: | 2018-07-05 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Serotonin transporter-ibogaine complexes illuminate mechanisms of inhibition and transport.
Nature, 569, 2019
|
|
6TT3
 
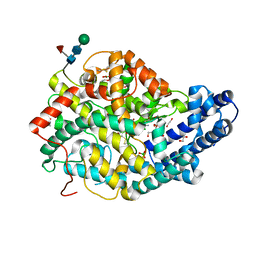 | |
8I9V
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - State Dbp10-2 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
8OM9
 
 | | MutSbeta bound to (CAG)2 DNA (open form) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | MutSbeta bound to (CAG)2 DNA (open form)
To Be Published
|
|
8I9W
 
 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - Dbp10-3 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
6QX6
 
 | | Structure of gtPebB-dihydrobiliverdin complex | | Descriptor: | 15,16-DIHYDROBILIVERDIN, Ferredoxin bilin reductase plastid, PENTAETHYLENE GLYCOL, ... | | Authors: | Sommerkamp, J.A, Hofmann, E. | | Deposit date: | 2019-03-07 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the first eukaryotic bilin reductaseGtPEBB reveals a flipped binding mode of dihydrobiliverdin.
J.Biol.Chem., 294, 2019
|
|
8ONO
 
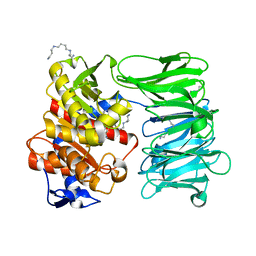 | | Modified oligopeptidase B from S. proteamaculans in intermediate conformation with 5 spermine molecule at 1.65 A resolution | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2023-04-03 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 5 spermine molecule at 1.65 A resolution
To Be Published
|
|
2X28
 
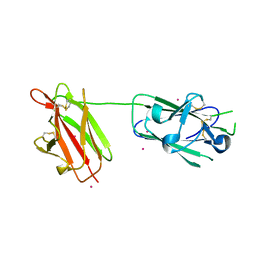 | | cadmium bound structure of SporoSAG | | Descriptor: | CADMIUM ION, SPOROZOITE-SPECIFIC SAG PROTEIN | | Authors: | Crawford, J, Lamb, E, Wasmuth, J, Grujic, O, Grigg, M.E, Boulanger, M.J. | | Deposit date: | 2010-01-11 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Characterization of Sporosag: A Sag2 Related Surface Antigen from Toxoplasma Gondii.
J.Biol.Chem., 285, 2010
|
|
7UUN
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with neomycin | | Descriptor: | 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, NEOMYCIN | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Osipiuk, J, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
8OLX
 
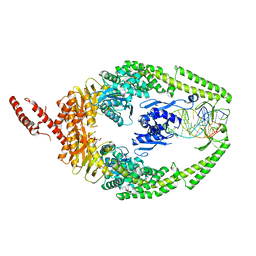 | | MutSbeta bound to (CAG)2 DNA (canonical form) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (25-MER), DNA mismatch repair protein Msh2, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-30 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | MutSbeta bound to (CAG)2 DNA (canonical form)
To Be Published
|
|
8I9Y
 
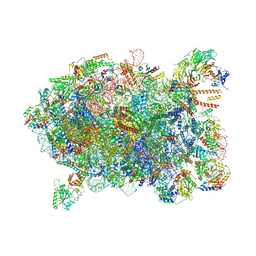 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - Ytm1-2 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
