2JJD
 
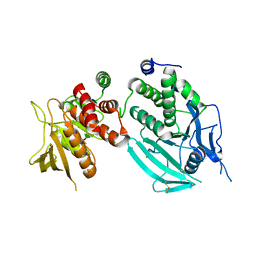 | | Protein Tyrosine Phosphatase, Receptor Type, E isoform | | Descriptor: | CHLORIDE ION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE EPSILON | | Authors: | Elkins, J.M, Ugochukwu, E, Alfano, I, Barr, A.J, Bunkoczi, G, King, O.N.F, Filippakopoulos, P, Savitsky, P, Salah, E, Pike, A, Johansson, C, Das, S, Burgess-Brown, N.A, Gileadi, O, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-08 | | Last modified: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Large-Scale Structural Analysis of the Classical Human Protein Tyrosine Phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2VCI
 
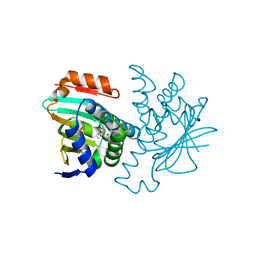 | | 4,5 Diaryl Isoxazole Hsp90 Chaperone Inhibitors: Potential Therapeutic Agents for the Treatment of Cancer | | Descriptor: | 5-[2,4-DIHYDROXY-5-(1-METHYLETHYL)PHENYL]-N-ETHYL-4-[4-(MORPHOLIN-4-YLMETHYL)PHENYL]ISOXAZOLE-3-CARBOXAMIDE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Brough, P.A, Aherne, W, Barril, X, Borgognoni, J, Boxal, K, Cansfield, J.E, Cheung, K.M, Collins, I, Davies, N.G.M, Drysdale, M.J, Dymock, B, Eccles, S.A, Finch, H, Fink, A, Hayes, A, Howes, R, Hubbard, R.E, James, K, Jordan, A.M, Lockie, A, Martins, V, Massey, A, Matthews, T.P, McDonald, E, Northfield, C.J, Pearl, L.H, Prodromou, C, Ray, S, Raynaud, F.I, Roughley, S.D, Sharp, S.Y, Surgenor, A, Walmsley, D.L, Webb, P, Wood, M, Workman, P, Wright, L. | | Deposit date: | 2007-09-24 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 4,5-diarylisoxazole Hsp90 chaperone inhibitors: potential therapeutic agents for the treatment of cancer.
J. Med. Chem., 51, 2008
|
|
7U6P
 
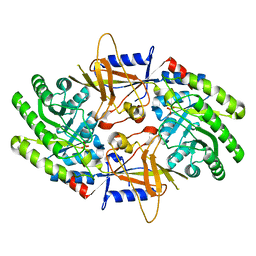 | | Structure of an intellectual disability-associated ornithine decarboxylase variant G84R | | Descriptor: | Ornithine decarboxylase, PHOSPHATE ION | | Authors: | Zhou, X.E, Schultz, C.R, Powell, K.S, Henrickson, A, Lamp, J, Brunzelle, J.S, Demeler, B, Vega, I.E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and Enzymatic Activity of an Intellectual Disability-Associated Ornithine Decarboxylase Variant, G84R.
Acs Omega, 7, 2022
|
|
7U6U
 
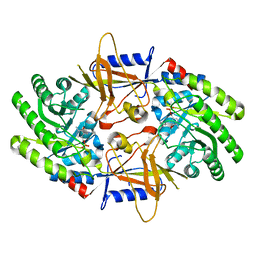 | | Structure of an intellectual disability-associated ornithine decarboxylase variant G84R in complex with PLP | | Descriptor: | Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Schultz, C.R, Powell, K.S, Henrickson, A, Lamp, J, Brunzelle, J.S, Demeler, B, Vega, I.E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2022-03-06 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Enzymatic Activity of an Intellectual Disability-Associated Ornithine Decarboxylase Variant, G84R.
Acs Omega, 7, 2022
|
|
5N8N
 
 | | Contracted sheath of a Pseudomonas aeruginosa type six secretion system consisting of TssB1 and TssC1 | | Descriptor: | EvpB family type VI secretion protein, Type VI secretion protein, family | | Authors: | Salih, O, He, S, Stach, L, Macdonald, J.T, Planamente, S, Manoli, E, Scheres, S, Filloux, A, Freemont, P.S. | | Deposit date: | 2017-02-23 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Atomic Structure of Type VI Contractile Sheath from Pseudomonas aeruginosa.
Structure, 26, 2018
|
|
6F2Q
 
 | | Neutron crystal structure of perdeuterated galectin-3C in the ligand-free form | | Descriptor: | Galectin-3 | | Authors: | Manzoni, F, Blakeley, M.P, Oksanen, E, Logan, D.T. | | Deposit date: | 2017-11-27 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.03 Å), X-RAY DIFFRACTION | | Cite: | Elucidation of Hydrogen Bonding Patterns in Ligand-Free, Lactose- and Glycerol-Bound Galectin-3C by Neutron Crystallography to Guide Drug Design.
J. Med. Chem., 61, 2018
|
|
7K8Q
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C121 | | Descriptor: | C121 Fab Heavy Chain, C121 Fab Light Chain, GLYCEROL | | Authors: | Abernathy, M.E, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
3IO9
 
 | | BimL12Y in complex with Mcl-1 | | Descriptor: | Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Czabotar, P.E, Lee, E.F, Yang, H, Sleebs, B.E, Lessene, G, Colman, P.M, Smith, B.J, Fairlie, W.D. | | Deposit date: | 2009-08-14 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands.
J.Biol.Chem., 284, 2009
|
|
3M88
 
 | | Crystal structure of the cysteine protease inhibitor, EhICP2, from Entamoeba histolytica | | Descriptor: | Amoebiasin-2, CHLORIDE ION, SULFATE ION | | Authors: | Lara-Gonzalez, S, Casados-Vazquez, L.E, Brieba, L.G. | | Deposit date: | 2010-03-17 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the cysteine protease inhibitor 2 from Entamoeba histolytica: functional convergence of a common protein fold.
Gene, 471, 2011
|
|
5V1M
 
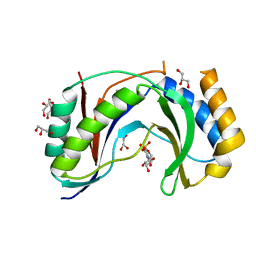 | | Structure of human Usb1 with uridine 5'-monophosphate | | Descriptor: | CHLORIDE ION, GLYCEROL, U6 snRNA phosphodiesterase, ... | | Authors: | Montemayor, E.J, Didychuk, A.L, Butcher, S.E. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Usb1 controls U6 snRNP assembly through evolutionarily divergent cyclic phosphodiesterase activities.
Nat Commun, 8, 2017
|
|
8CIH
 
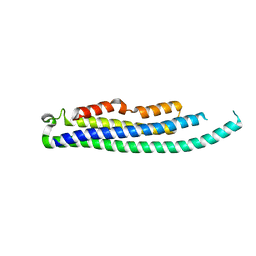 | | Structure of FL CINP | | Descriptor: | Cyclin-dependent kinase 2-interacting protein, SODIUM ION | | Authors: | Foglizzo, M, Zeqiraj, E. | | Deposit date: | 2023-02-09 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SPATA5-SPATA5L1 ATPase complex directs replisome proteostasis to ensure genome integrity.
Cell, 187, 2024
|
|
1BJX
 
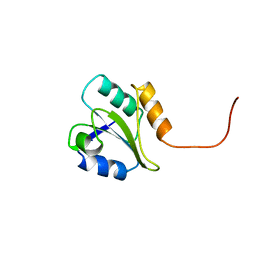 | | HUMAN PROTEIN DISULFIDE ISOMERASE, NMR, 24 STRUCTURES | | Descriptor: | PROTEIN DISULFIDE ISOMERASE | | Authors: | Kemmink, J, Dijkstra, K, Mariani, M, Scheek, R.M, Penka, E, Nilges, M, Darby, N.J. | | Deposit date: | 1998-06-29 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure in solution of the b domain of protein disulfide isomerase.
J.Biomol.NMR, 13, 1999
|
|
6F8X
 
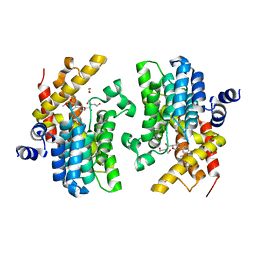 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-26g | | Descriptor: | 1,2-ETHANEDIOL, 2-[(5~{R})-3-(3-cyclopentyloxy-4-methoxy-phenyl)-4,5-dihydro-1,2-oxazol-5-yl]-~{N},~{N}-bis(2-hydroxyethyl)ethanamide, DIMETHYL SULFOXIDE, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
5FIF
 
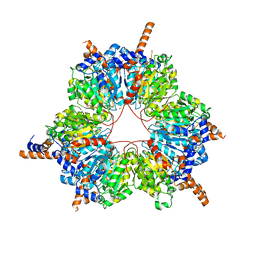 | | Carboxyltransferase domain of a single-chain bacterial carboxylase | | Descriptor: | 1,2-ETHANEDIOL, Carboxylase | | Authors: | Hagmann, A, Hunkeler, M, Stuttfeld, E, Maier, T. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Hybrid Structure of a Dynamic Single-Chain Carboxylase from Deinococcus radiodurans.
Structure, 24, 2016
|
|
8C5I
 
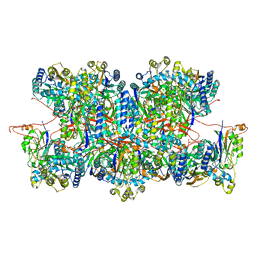 | | Cyanide dihydratase from Bacillus pumilus C1 variant - Q86R,H305K,H308K,H323K | | Descriptor: | Cyanide dihydratase | | Authors: | Mulelu, A.E, Reitz, J, van Rooyen, J, Scheffer, M, Frangakis, A.S, Dlamini, L.S, Woodward, J.D, Benedik, M.J, Sewell, B.T. | | Deposit date: | 2023-01-09 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | The Role of Histidine Residues in the Oligomerization of Cyanide Dihydratase from Bacillus pumilus C1
To Be Published
|
|
1NOV
 
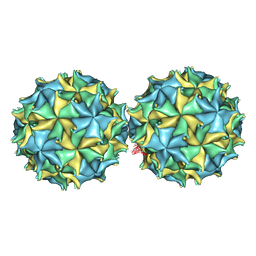 | | NODAMURA VIRUS | | Descriptor: | NODAMURA VIRUS COAT PROTEINS | | Authors: | Natarajan, P, Johnson, J.E. | | Deposit date: | 1997-09-16 | | Release date: | 1998-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Resolution of space-group ambiguity and structure determination of nodamura virus to 3.3 A resolution from pseudo-R32 (monoclinic) crystals.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
6PS5
 
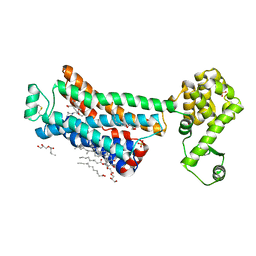 | | XFEL beta2 AR structure by ligand exchange from Timolol to Propranolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(ISOPROPYLAMINO)-3-(1-NAPHTHYLOXY)-2-PROPANOL, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
6QKU
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Tyr219Phe - Chloroacetate soaked 2hr | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, GLYCOLIC ACID, ... | | Authors: | Mehrabi, P, Kim, T.H, Prosser, R.S, Pai, E.F. | | Deposit date: | 2019-01-30 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | Substrate-Based Allosteric Regulation of a Homodimeric Enzyme.
J.Am.Chem.Soc., 141, 2019
|
|
6F9T
 
 | | Crystal structure of human testis Angiotensin-1 converting enzyme in complex with Sampatrilat. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of sampatrilat and sampatrilat-Asp in complex with human ACE - a molecular basis for domain selectivity.
FEBS J., 285, 2018
|
|
7TS0
 
 | | Cryo-EM structure of corticotropin releasing factor receptor 2 bound to Urocortin 1 and coupled with heterotrimeric Go protein | | Descriptor: | Corticotropin-releasing factor receptor 2,Corticotropin-releasing factor receptor 2,Corticotropin-releasing factor receptor 2,Corticotropin-releasing factor receptor 2,Human corticotropin releasing factor receptor 2, Dominant negative Go alpha subunit, G protein gamma subunit, ... | | Authors: | Zhao, L.-H, Lin, J, Mao, C, Zhou, X.E, Ji, S, Shen, D, Xiao, P, Melcher, K, Zhang, Y, Yu, X, Xu, H.E. | | Deposit date: | 2022-01-31 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure insights into selective coupling of G protein subtypes by a class B G protein-coupled receptor.
Nat Commun, 13, 2022
|
|
7TRY
 
 | | Cryo-EM structure of corticotropin releasing factor receptor 2 bound to Urocortin 1 and coupled with heterotrimeric G11 protein | | Descriptor: | Corticotropin-releasing factor receptor 2, G protein gamma subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, L.-H, Lin, J, Mao, C, Zhou, X.E, Ji, S, Shen, D, Xiao, P, Melcher, K, Zhang, Y, Yu, X, Xu, H.E. | | Deposit date: | 2022-01-31 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure insights into selective coupling of G protein subtypes by a class B G protein-coupled receptor.
Nat Commun, 13, 2022
|
|
8RAI
 
 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis point mutant R90I complexed with phenylhydrazine | | Descriptor: | Aminotransferase class IV, GLYCEROL, [6-methyl-5-oxidanyl-4-[(2-phenylhydrazinyl)methyl]pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y, Boyko, K.M. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multifunctionality of arginine residues in the active sites of non-canonical d-amino acid transaminases.
Arch.Biochem.Biophys., 756, 2024
|
|
3M3J
 
 | | A new crystal form of Lys48-linked diubiquitin | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Ubiquitin | | Authors: | Trempe, J.F, Brown, N.R, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new crystal form of Lys48-linked diubiquitin.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2W36
 
 | | Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*TP*GP*TP*AP*GP*CP)-3', 5'-D(*GP*CP*BRUP*AP*CP*IP*GP*AP*BRUP*CP*GP)-3', ENDONUCLEASE V | | Authors: | Dalhus, B, Arvai, A.S, Rosnes, I, Olsen, O.E, Backe, P.H, Alseth, I, Gao, H, Cao, W, Tainer, J.A, Bjoras, M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Endonuclease V with DNA Reveal Initiation of Deaminated Adenine Repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
8RAF
 
 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis point mutant R90I (holo form) | | Descriptor: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y, Boyko, K.M. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multifunctionality of arginine residues in the active sites of non-canonical d-amino acid transaminases.
Arch.Biochem.Biophys., 756, 2024
|
|
