2AAF
 
 | |
1O08
 
 | | Structure of Pentavalent Phosphorous Intermediate of an Enzyme Catalyzed Phosphoryl transfer Reaction observed on cocrystallization with Glucose 1-phosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-02-20 | | Release date: | 2003-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The pentacovalent phosphorus intermediate of a phosphoryl transfer reaction.
Science, 299, 2003
|
|
2ABR
 
 | |
2A9G
 
 | | Structure of C406A arginine deiminase in complex with L-arginine | | Descriptor: | ARGININE, Arginine deiminase | | Authors: | Galkin, A, Lu, X, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2005-07-11 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures Representing the Michaelis Complex and the Thiouronium Reaction Intermediate of Pseudomonas aeruginosa Arginine Deiminase.
J.Biol.Chem., 280, 2005
|
|
2ACI
 
 | |
4K4A
 
 | | X-ray crystal structure of E. coli YdiI complexed with phenacyl-CoA | | Descriptor: | Esterase YdiI, phenacyl coenzyme A | | Authors: | Ru, W, Farelli, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2013-04-12 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and Catalysis in the Escherichia coli Hotdog-fold Thioesterase Paralogs YdiI and YbdB.
Biochemistry, 53, 2014
|
|
4K4D
 
 | | X-ray crystal structure of E. coli YbdB complexed with 2,4-dihydroxyphenacyl-CoA | | Descriptor: | 2,4-dihydroxyphenacyl coenzyme A, ACETATE ION, MALONATE ION, ... | | Authors: | Ru, W, Farelli, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2013-04-12 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and Catalysis in the Escherichia coli Hotdog-fold Thioesterase Paralogs YdiI and YbdB.
Biochemistry, 53, 2014
|
|
4K4B
 
 | | X-ray crystal structure of E. coli YdiI complexed with undeca-2-one-CoA | | Descriptor: | CHLORIDE ION, Esterase YdiI, undeca-2-one coenzyme A | | Authors: | Ru, W, Farelli, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2013-04-12 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Catalysis in the Escherichia coli Hotdog-fold Thioesterase Paralogs YdiI and YbdB.
Biochemistry, 53, 2014
|
|
4K4C
 
 | | X-ray crystal structure of E. coli YbdB complexed with phenacyl-CoA | | Descriptor: | Proofreading thioesterase EntH, phenacyl coenzyme A | | Authors: | Ru, W, Farelli, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2013-04-12 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Catalysis in the Escherichia coli Hotdog-fold Thioesterase Paralogs YdiI and YbdB.
Biochemistry, 53, 2014
|
|
4K49
 
 | | X-ray crystal structure of E. coli YdiI complexed with 2,4-dihydroxyphenacyl CoA | | Descriptor: | 2,4-dihydroxyphenacyl coenzyme A, Esterase YdiI | | Authors: | Ru, W, Farelli, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2013-04-12 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and Catalysis in the Escherichia coli Hotdog-fold Thioesterase Paralogs YdiI and YbdB.
Biochemistry, 53, 2014
|
|
3DLP
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, Mutant D402P, bound to 4CB | | Descriptor: | 4-CHLORO-BENZOIC ACID, 4-Chlorobenzoate CoA Ligase/Synthetase | | Authors: | Wu, R, Cao, J, Reger, A.S, Lu, X, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2008-06-28 | | Release date: | 2009-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The mechanism of domain alternation in the acyl-adenylate forming ligase superfamily member 4-chlorobenzoate: coenzyme A ligase
Biochemistry, 48, 2009
|
|
3E8M
 
 | | Structure-function Analysis of 2-Keto-3-deoxy-D-glycero-D-galacto-nononate-9-phosphate (KDN) Phosphatase Defines a New Clad Within the Type C0 HAD Subfamily | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Acylneuraminate cytidylyltransferase, ... | | Authors: | Lu, Z, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2008-08-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure-Function Analysis of 2-Keto-3-deoxy-D-glycero-D-galactonononate-9-phosphate Phosphatase Defines Specificity Elements in Type C0 Haloalkanoate Dehalogenase Family Members.
J.Biol.Chem., 284, 2009
|
|
4JB3
 
 | | Crystal structure of BT_0970, a had family phosphatase from bacteroides thetaiotaomicron VPI-5482, TARGET EFI-501083, with bound sodium and glycerol, closed lid, ordered loop | | Descriptor: | GLYCEROL, Haloacid dehalogenase-like hydrolase, SODIUM ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Kumar, P.R, Ghosh, A, Al Obaidi, N.F, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of bt_0970, a had family phosphatase from bacteroides thetaiotaomicron VPI-5482, TARGET EFI-501083, with bound sodium and glycerol, closed lid, ordered loop
To be Published
|
|
4DWO
 
 | | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Mg crystal form II | | Descriptor: | GLYCEROL, Haloacid dehalogenase-like hydrolase, MAGNESIUM ION | | Authors: | Vetting, M.W, Wasserman, S.R, Morisco, L.L, Sojitra, S, Allen, K.N, Dunaway-Mariano, D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-26 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Mg crystal form II
To be Published
|
|
4DW8
 
 | | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Na crystal form I | | Descriptor: | Haloacid dehalogenase-like hydrolase, SODIUM ION, UNKNOWN LIGAND | | Authors: | Vetting, M.W, Wasserman, S.R, Morisco, L.L, Sojitra, S, Allen, K.N, Dunaway-Mariano, D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-24 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Na crystal form I
To be Published
|
|
4G9B
 
 | | Crystal structure of beta-phosphoglucomutase homolog from escherichia coli, target efi-501172, with bound mg, open lid | | Descriptor: | Beta-phosphoglucomutase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of beta-phosphoglucomutase homolog from escherichia coli, target efi-501172, with bound mg, open lid
To be Published
|
|
1OQF
 
 | | Crystal structure of the 2-methylisocitrate lyase | | Descriptor: | 2-methylisocitrate lyase | | Authors: | Liu, S, Lu, Z, Dunaway-Mariano, D, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-03-08 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of 2-methylisocitrate lyase in complex with product and with isocitrate inhibitor provide insight into lyase substrate specificity, catalysis and evolution.
Biochemistry, 44, 2005
|
|
1Q4S
 
 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase complexed with CoA and 4-hydroxybenzoic acid | | Descriptor: | COENZYME A, P-HYDROXYBENZOIC ACID, Thioesterase | | Authors: | Thoden, J.B, Zhuang, Z, Dunaway-Mariano, D, Holden, H.M. | | Deposit date: | 2003-08-04 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Structure of 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. strain SU
J.Biol.Chem., 278, 2003
|
|
1Q4U
 
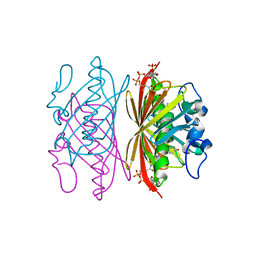 | | Crystal structure of 4-hydroxybenzoyl CoA thioesterase from arthrobacter sp. strain SU complexed with 4-hydroxybenzyl CoA | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXYBENZYL COENZYME A, Thioesterase | | Authors: | Thoden, J.B, Zhuang, Z, Dunaway-Mariano, D, Holden, H.M. | | Deposit date: | 2003-08-04 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. strain SU
J.Biol.Chem., 278, 2003
|
|
1Q4T
 
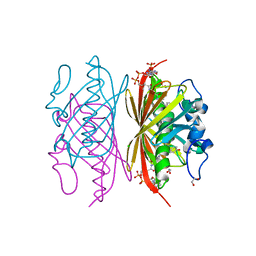 | | crystal structure of 4-hydroxybenzoyl CoA thioesterase from Arthrobacter sp. strain SU complexed with 4-hydroxyphenyl CoA | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXYPHENACYL COENZYME A, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Zhuang, Z, Dunaway-Mariano, D, Holden, H.M. | | Deposit date: | 2003-08-04 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. strain SU
J.Biol.Chem., 278, 2003
|
|
1RQL
 
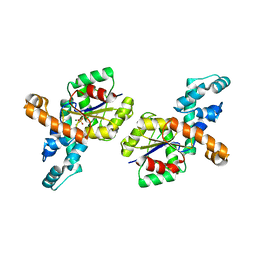 | | Crystal Structure of Phosponoacetaldehyde Hydrolase Complexed with Magnesium and the Inhibitor Vinyl Sulfonate | | Descriptor: | MAGNESIUM ION, Phosphonoacetaldehyde Hydrolase, VINYLSULPHONIC ACID | | Authors: | Morais, M.C, Zhang, G, Zhang, W, Olsen, D.B, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-12-05 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic and site-directed mutagenesis
analysis of the mechanism of Schiff-base formation in
phosphonoacetaldehyde hydrolase catalysis
J.Biol.Chem., 279, 2004
|
|
1RDF
 
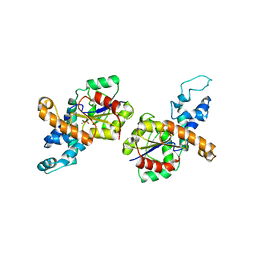 | | G50P mutant of phosphonoacetaldehyde hydrolase in complex with substrate analogue vinyl sulfonate | | Descriptor: | ETHANESULFONIC ACID, MAGNESIUM ION, phosphonoacetaldehyde hydrolase | | Authors: | Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-11-05 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the substrate specificity loop of the HAD superfamily cap domain
Biochemistry, 43, 2004
|
|
1RQN
 
 | | Phosphonoacetaldehyde hydrolase complexed with magnesium | | Descriptor: | MAGNESIUM ION, Phosphonoacetaldehyde Hydrolase | | Authors: | Morais, M.C, Zhang, G, Zhang, W, Olsen, D.B, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-12-05 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic and site-directed mutagenesis
analysis of the mechanism of Schiff-base formation in
phosphonoacetaldehyde hydrolase catalysis
J.Biol.Chem., 279, 2004
|
|
1S2W
 
 | | Crystal structure of phosphoenolpyruvate mutase in high ionic strength | | Descriptor: | Phosphoenolpyruvate phosphomutase, SULFATE ION | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1S2T
 
 | | Crystal Structure Of Apo Phosphoenolpyruvate Mutase | | Descriptor: | Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
