5HFS
 
 | | CRYSTAL STRUCTURE OF C-TERMINAL DOMAIN OF CARGO PROTEINS OF TYPE IX SECRETION SYSTEM | | 分子名称: | CALCIUM ION, Gingipain R2, ZINC ION | | 著者 | Golik, P, Szmigielski, B, Ksiazek, M, Nowakowska, Z, Mizgalska, D, Nowak, M, Dubin, G, Potempa, J. | | 登録日 | 2016-01-07 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal beta-sandwich domain.
Sci Rep, 6, 2016
|
|
5FHT
 
 | | HtrA2 protease mutant V226K | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, POTASSIUM ION, ... | | 著者 | Golik, P, Dubin, G, Zurawa-Janicka, D, Lipinska, B, Jarzab, M, Wenta, T, Gieldon, A, Ciarkowski, A. | | 登録日 | 2015-12-22 | | 公開日 | 2016-09-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Distinct 3D Architecture and Dynamics of the Human HtrA2(Omi) Protease and Its Mutated Variants.
Plos One, 11, 2016
|
|
5J7F
 
 | | Structure of MDM2 with low molecular weight inhibitor with aliphatic linker. | | 分子名称: | 4-({6-[(6-chloro-3-{1-[(4-chlorophenyl)methyl]-4-(4-fluorophenyl)-1H-imidazol-5-yl}-1H-indole-2-carbonyl)oxy]hexyl}amino)-4-oxobutanoic acid, E3 ubiquitin-protein ligase Mdm2 | | 著者 | Twarda-Clapa, A, Kubica, K, Guzik, K, Dubin, G, Holak, T.A. | | 登録日 | 2016-04-06 | | 公開日 | 2017-05-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | 1,4,5-Trisubstituted Imidazole-Based p53-MDM2/MDMX Antagonists with Aliphatic Linkers for Conjugation with Biological Carriers.
J. Med. Chem., 60, 2017
|
|
4TRP
 
 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | 分子名称: | Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen | | 著者 | Horwacik, I, Golik, P, Grudnik, P, Zdzalik, M, Rokita, H, Dubin, G. | | 登録日 | 2014-06-17 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
4U2W
 
 | | Atomic resolution crystal structure of HV-BBI protease inhibitor from amphibian skin in complex with bovine trypsin | | 分子名称: | 1,2-ETHANEDIOL, Bowman-Birk trypsin inhibitor, CALCIUM ION, ... | | 著者 | Grudnik, P, Golik, P, Malicki, S, Debowski, D, Legowska, A, Rolka, K, Dubin, G. | | 登録日 | 2014-07-18 | | 公開日 | 2015-01-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Atomic resolution crystal structure of HV-BBI protease inhibitor from amphibian skin in complex with bovine trypsin.
Proteins, 83, 2015
|
|
4TUO
 
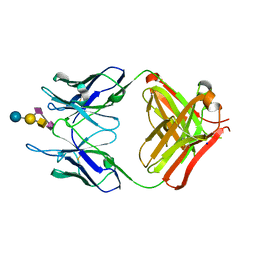 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | 分子名称: | Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-[2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)]beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | 著者 | Golik, P, Grudnik, P, Horwacik, I, Zdzalik, M, Rokita, H, Dubin, G. | | 登録日 | 2014-06-24 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
4TUL
 
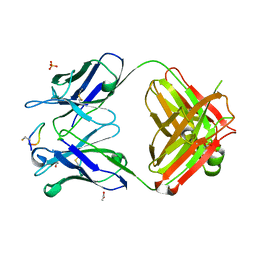 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | 分子名称: | ACETATE ION, Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen, ... | | 著者 | Golik, P, Grudnik, P, Dubin, G, Zdzalik, M, Rokita, H, Horwacik, I. | | 登録日 | 2014-06-24 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
4TUJ
 
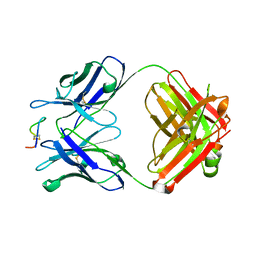 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | 分子名称: | Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen, peptide1 | | 著者 | Grudnik, P, Golik, P, Horwacik, I, Zdzalik, M, Rokita, H, Dubin, G. | | 登録日 | 2014-06-24 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
4TUK
 
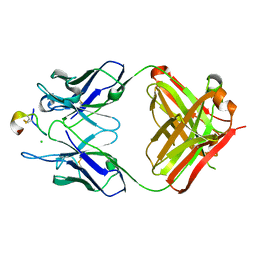 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | 分子名称: | CHLORIDE ION, Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen, ... | | 著者 | Golik, P, Grudnik, P, Dubin, G, Horwacik, I, Zdzalik, M, Rokita, H. | | 登録日 | 2014-06-24 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
4ZFI
 
 | | Structure of Mdm2 with low molecular weight inhibitor | | 分子名称: | (5S)-3,5-bis(4-chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)-5-hydroxy-1-methyl-1,5-dihydro-2H-pyrrol-2-one, E3 ubiquitin-protein ligase Mdm2 | | 著者 | Zak, K.M, Twarda-Clapa, A, Wrona, E.M, Grudnik, P, Dubin, G, Holak, T.A. | | 登録日 | 2015-04-21 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction.
ACS Chem. Biol., 11, 2016
|
|
4ZGK
 
 | | Structure of Mdm2 with low molecular weight inhibitor. | | 分子名称: | (5R)-3,5-bis(4-chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)-5-hydroxyfuran-2(5H)-one, E3 ubiquitin-protein ligase Mdm2 | | 著者 | Twarda-Clapa, A, Zak, K.M, Wrona, E.M, Grudnik, P, Dubin, G, Holak, T.A. | | 登録日 | 2015-04-23 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction.
ACS Chem. Biol., 11, 2016
|
|
4XOJ
 
 | | Structure of bovine trypsin in complex with analogues of sunflower inhibitor 1 (SFTI-1) | | 分子名称: | 1,2-ETHANEDIOL, AMMONIUM ION, CALCIUM ION, ... | | 著者 | Golik, P, Malicki, S, Grudnik, P, Karna, N, Debowski, D, Legowska, A, Wladyka, B, Gitlin, A, Brzozowski, K, Dubin, G, Rolka, K. | | 登録日 | 2015-01-16 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (0.91 Å) | | 主引用文献 | Investigation of Serine-Proteinase-Catalyzed Peptide Splicing in Analogues of Sunflower Trypsin Inhibitor 1 (SFTI-1).
Chembiochem, 16, 2015
|
|
4ZQK
 
 | | Structure of the complex of human programmed death-1 (PD-1) and its ligand PD-L1. | | 分子名称: | Programmed cell death 1 ligand 1, Programmed cell death protein 1, SODIUM ION | | 著者 | Zak, K.M, Dubin, G, Holak, T.A. | | 登録日 | 2015-05-10 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structure of the Complex of Human Programmed Death 1, PD-1, and Its Ligand PD-L1.
Structure, 23, 2015
|
|
5AG8
 
 | | CRYSTAL STRUCTURE OF A MUTANT (665I6H) OF THE C-TERMINAL DOMAIN OF RGPB | | 分子名称: | GINGIPAIN R2, GLYCEROL, SULFATE ION | | 著者 | de Diego, I, Ksiazek, M, Mizgalska, D, Golik, P, Szmigielski, B, Nowak, M, Nowakowska, Z, Potempa, B, Koneru, L, Nguyen, K.A, Enghild, J, Thogersen, I.B, Dubin, G, Gomis-Ruth, F.X, Potempa, J. | | 登録日 | 2015-01-29 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The Outer-Membrane Export Signal of Porphyromonas Gingivalis Type Ix Secretion System (T9Ss) is a Conserved C-Terminal Beta-Sandwich Domain.
Sci.Rep., 6, 2016
|
|
5AG9
 
 | | CRYSTAL STRUCTURE OF A MUTANT (665sXa) C-TERMINAL DOMAIN OF RGPB | | 分子名称: | Gingipain R2, SULFATE ION | | 著者 | de Diego, I, Ksiazek, M, Mizgalska, D, Golik, P, Szmigielski, B, Nowak, M, Nowakowska, Z, Potempa, B, Koneru, L, Nguyen, K.A, Enghild, J, Thogersen, I.B, Dubin, G, Gomis-Ruth, F.X, Potempa, J. | | 登録日 | 2015-01-29 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal beta-sandwich domain.
Sci Rep, 6, 2016
|
|
5OML
 
 | | Crystal structure of Trypanosoma Brucei PEX14 N-terminal domain in complex with small molecules to investigate the water envelope | | 分子名称: | (3~{R})-3-[[1-(2-hydroxyethyl)-5-[(4-methoxynaphthalen-1-yl)methyl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-3-yl]carbonylamino]-3-phenyl-propanoic acid, BETA-MERCAPTOETHANOL, Peroxin 14, ... | | 著者 | Ratkova, E.L, Dawidowski, M, Napolitano, V, Dubin, G, Fino, R, Popowicz, G, Sattler, M, Tetko, I.V. | | 登録日 | 2017-08-01 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of Trypanosoma Brucei PEX14 N-terminal domain in complex with small molecules to investigate the water envelope
To Be Published
|
|
5N4K
 
 | | N-terminal domain of a human Coronavirus NL63 nucleocapsid protein | | 分子名称: | Nucleoprotein, SULFATE ION | | 著者 | Zdzalik, M, Szelazek, B, Kabala, W, Golik, P, Burmistrz, M, Florek, D, Kus, K, Pyrc, K, Dubin, G. | | 登録日 | 2017-02-10 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structural Characterization of Human Coronavirus NL63 N Protein.
J. Virol., 91, 2017
|
|
5N2F
 
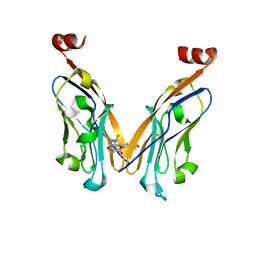 | | Structure of PD-L1/small-molecule inhibitor complex | | 分子名称: | 4-[[4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]-2,5-bis(fluoranyl)phenyl]methylamino]-3-oxidanylidene-butanoic acid, Programmed cell death 1 ligand 1 | | 著者 | Guzik, K, Zak, K.M, Grudnik, P, Dubin, G, Holak, T.A. | | 登録日 | 2017-02-07 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) Interaction via Transiently Induced Protein States and Dimerization of PD-L1.
J. Med. Chem., 60, 2017
|
|
5N86
 
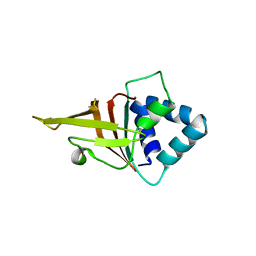 | | Crystal structure of FAS1 domain of hyaluronic acid receptor stabilin-2 | | 分子名称: | Stabilin-2 | | 著者 | Twarda-Clapa, A, Labuzek, B, Grudnik, P, Dubin, G, Holak, T.A. | | 登録日 | 2017-02-23 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.484 Å) | | 主引用文献 | Crystal structure of the FAS1 domain of the hyaluronic acid receptor stabilin-2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5NIF
 
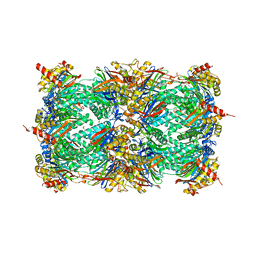 | | Yeast 20S proteasome in complex with Blm-pep activator | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Witkowska, J, Grudnik, P, Golik, P, Dubin, G, Jankowska, E. | | 登録日 | 2017-03-23 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of a low molecular weight activator Blm-pep with yeast 20S proteasome - insights into the enzyme activation mechanism.
Sci Rep, 7, 2017
|
|
5NM7
 
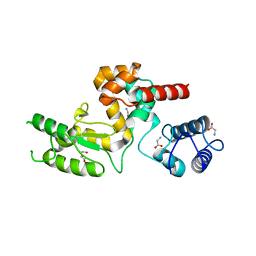 | | Crystal structure of Burkholderia AP3 phage endolysin | | 分子名称: | GLYCINE, Peptidoglycan-binding domain 1, TRIETHYLENE GLYCOL | | 著者 | Zrubek, K, Wisniewska, M, Rembacz, K, Maciejewska, B, Drulis-Kawa, Z, Dubin, G. | | 登録日 | 2017-04-05 | | 公開日 | 2018-02-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Modular endolysin of Burkholderia AP3 phage has the largest lysozyme-like catalytic subunit discovered to date and no catalytic aspartate residue.
Sci Rep, 7, 2017
|
|
5O0I
 
 | |
5O4Y
 
 | | Structure of human PD-L1 in complex with inhibitor | | 分子名称: | PHE-MAA-ASN-PRO-HIS-LEU-SER-TRP-SER-TRP-9KK-9KK-ARG-CCS-GLY-NH2, Programmed cell death 1 ligand 1 | | 著者 | Magiera, K, Grudnik, P, Dubin, G, Holak, T.A. | | 登録日 | 2017-05-31 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Bioactive Macrocyclic Inhibitors of the PD-1/PD-L1 Immune Checkpoint.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MM8
 
 | |
5OAI
 
 | | Structure of MDM2 with low molecular weight inhibitor | | 分子名称: | 3-[(1~{R})-2-(~{tert}-butylamino)-1-[methanoyl-[[3,4,5-tris(fluoranyl)phenyl]methyl]amino]-2-oxidanylidene-ethyl]-6-chloranyl-1~{H}-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2 | | 著者 | Twarda-Clapa, A, Neochoritis, C.G, Grudnik, P, Dubin, G, Domling, A, Holak, T.A. | | 登録日 | 2017-06-22 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A fluorinated indole-based MDM2 antagonist selectively inhibits the growth of p53wtosteosarcoma cells.
Febs J., 286, 2019
|
|
