5Y0V
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with berberine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BERBERINE, ... | | Authors: | Duan, Y.W, Liu, T, Tang, J.Y, Li, M, Yang, Q. | | Deposit date: | 2017-07-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | Glycoside hydrolase family 18 and 20 enzymes are novel targets of the traditional medicine berberine.
J. Biol. Chem., 293, 2018
|
|
5Y1B
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with a berberine derivative (SYSU-00679) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-3'-quinolinium propylberberine, Beta-hexosaminidase | | Authors: | Duan, Y.W, Liu, T, Zhou, Y, Tang, J.Y, Li, M, Yang, Q. | | Deposit date: | 2017-07-20 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.207 Å) | | Cite: | Glycoside hydrolase family 18 and 20 enzymes are novel targets of the traditional medicine berberine.
J. Biol. Chem., 293, 2018
|
|
5XP5
 
 | | C-Src in complex with ATP-Chf | | Descriptor: | MAGNESIUM ION, Proto-oncogene tyrosine-protein kinase Src, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-[(S)-fluoranyl-[oxidanyl(phosphonooxy)phosphoryl]methyl]phosphinic acid | | Authors: | Duan, Y, Guo, M, Dai, S, Chen, L, Chen, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Remarkably Stereospecific Utilization of ATP alpha , beta-Halomethylene Analogues by Protein Kinases.
J. Am. Chem. Soc., 139, 2017
|
|
7VUA
 
 | | Anaerobic hydroxyproline degradation involving C-N cleavage by a glycyl radical enzyme | | Descriptor: | (4S)-4-hydroxy-D-proline, HplG | | Authors: | Duan, Y, Lu, Q, Yuchi, Z, Zhang, Y. | | Deposit date: | 2021-11-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Anaerobic Hydroxyproline Degradation Involving C-N Cleavage by a Glycyl Radical Enzyme.
J.Am.Chem.Soc., 144, 2022
|
|
7DOV
 
 | |
5XP7
 
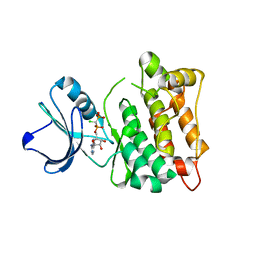 | | C-Src in complex with ATP-CHCl | | Descriptor: | GLYCEROL, MAGNESIUM ION, Proto-oncogene tyrosine-protein kinase Src, ... | | Authors: | Guo, M, Dai, S, Duan, Y, Chen, L, Chen, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Remarkably Stereospecific Utilization of ATP alpha , beta-Halomethylene Analogues by Protein Kinases.
J. Am. Chem. Soc., 139, 2017
|
|
4U5J
 
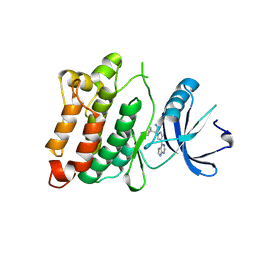 | | C-Src in complex with Ruxolitinib | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Chen, Y, Duan, Y, Chen, L. | | Deposit date: | 2014-07-25 | | Release date: | 2014-09-17 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | c-Src Binds to the Cancer Drug Ruxolitinib with an Active Conformation
Plos One, 9, 2014
|
|
9ITB
 
 | | LPA-bound LPAR6 in complex with miniGq | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2024-07-19 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Molecular mechanism of ligand recognition and activation of lysophosphatidic acid receptor LPAR6.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITE
 
 | | LPA-bound LPAR6 in complex with miniG13 | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2024-07-19 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Molecular mechanism of ligand recognition and activation of lysophosphatidic acid receptor LPAR6.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
4FPD
 
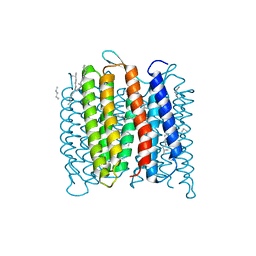 | | Deprotonation of D96 in bacteriorhodopsin opens the proton uptake pathway | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, CHLORIDE ION, ... | | Authors: | Wang, T, Sessions, A.O, Lunde, C.S, Rouani, S, Glaeser, R.M, Facciotti, M.T, Duan, Y. | | Deposit date: | 2012-06-22 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Deprotonation of d96 in bacteriorhodopsin opens the proton uptake pathway.
Structure, 21, 2013
|
|
7WF5
 
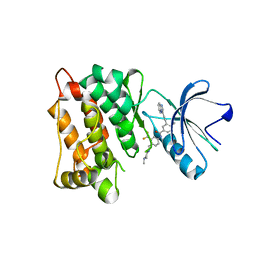 | | c-Src in complex with ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Guo, M, Duan, Y, Dai, S, Chen, X, Chen, Y. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural study of ponatinib in inhibiting SRC kinase.
Biochem.Biophys.Res.Commun., 598, 2022
|
|
5Z0V
 
 | | Structural insight into the Zika virus capsid encapsulating the viral genome | | Descriptor: | Extracellular solute-binding protein family 1,viral genome protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, T, Zhao, Q, Yang, X, Chen, C, Yang, K, Wu, C, Zhang, T, Duan, Y, Xue, X, Mi, K, Ji, X, Wang, Z, Yang, H. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structural insight into the Zika virus capsid encapsulating the viral genome.
Cell Res., 28, 2018
|
|
8WPH
 
 | | Anabaena McyI with prebound NAD | | Descriptor: | McyI-NAD, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, X, Yin, Y, Duan, Y, Liu, L. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the catalytic mechanism of the microcystin tailoring enzyme McyI.
Commun Biol, 8, 2025
|
|
8WPJ
 
 | | Anabaena McyI with prebound NAD and soaked NAD | | Descriptor: | McyI, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, X, Yin, Y, Duan, Y, Liu, L. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the catalytic mechanism of the microcystin tailoring enzyme McyI.
Commun Biol, 8, 2025
|
|
8WPI
 
 | | Anabaena McyI with prebound NAD and soaked NADP | | Descriptor: | McyI, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, X, Yin, Y, Duan, Y, Liu, L. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural insights into the catalytic mechanism of the microcystin tailoring enzyme McyI.
Commun Biol, 8, 2025
|
|
5Z0R
 
 | | Structural insight into the Zika virus capsid encapsulating the viral genome | | Descriptor: | Extracellular solute-binding protein family 1,viral genome protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, T, Zhao, Q, Yang, X, Chen, C, Yang, K, Wu, C, Zhang, T, Duan, Y, Xue, X, Mi, K, Ji, X, Wang, Z, Yang, H. | | Deposit date: | 2017-12-20 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insight into the Zika virus capsid encapsulating the viral genome.
Cell Res., 28, 2018
|
|
8WPQ
 
 | | Anabaena McyI R166M with prebound NAD and citrate | | Descriptor: | CITRATE ANION, McyI, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, X, Yin, Y, Duan, Y, Liu, L. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the catalytic mechanism of the microcystin tailoring enzyme McyI.
Commun Biol, 8, 2025
|
|
8WPS
 
 | | Anabaena McyI R166M with prebound NAD and malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, McyI, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, X, Yin, Y, Duan, Y, Liu, L. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the catalytic mechanism of the microcystin tailoring enzyme McyI.
Commun Biol, 8, 2025
|
|
8WPO
 
 | | Anabaena McyI R166A with prebound NAD and citrate | | Descriptor: | CITRATE ANION, McyI, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Wang, X, Yin, Y, Duan, Y, Liu, L. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the catalytic mechanism of the microcystin tailoring enzyme McyI.
Commun Biol, 8, 2025
|
|
8WPR
 
 | | Anabaena McyI R166A with prebound NAD and malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, GLYCEROL, McyI, ... | | Authors: | Wang, X, Yin, Y, Duan, Y, Liu, L. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic mechanism of the microcystin tailoring enzyme McyI.
Commun Biol, 8, 2025
|
|
5ZUM
 
 | | Structure of dipeptidyl-peptidase III from Corallococcus sp. strain EGB | | Descriptor: | ZINC ION, dipeptidyl-peptidase III | | Authors: | Zhang, H, Duan, Y.J, Li, Z.K, Liu, W.D, Huang, Y, Cui, Z.L. | | Deposit date: | 2018-05-08 | | Release date: | 2019-06-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of dipeptidyl peptidase III from Corallococcus sp. strain EGB
To Be Published
|
|
6AIN
 
 | | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PnpA | | Authors: | Chen, Q.Z, Huang, Y, Duan, Y.J, Li, Z.K, Liu, W.D, Cui, Z.L. | | Deposit date: | 2018-08-24 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4: The key enzyme involved in p-nitrophenol degradation.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
6AIO
 
 | | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4 | | Descriptor: | PnpA | | Authors: | Chen, Q.Z, Huang, Y, Duan, Y.J, Li, Z.K, Liu, W.D, Cui, Z.L. | | Deposit date: | 2018-08-24 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4: The key enzyme involved in p-nitrophenol degradation.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
7Y0F
 
 | | Crystal structure of TMPRSS2 in complex with UK-371804 | | Descriptor: | 2-[(1-carbamimidamido-4-chloranyl-isoquinolin-7-yl)sulfonylamino]-2-methyl-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, H, Duan, Y, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-06-04 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
7Y0E
 
 | | Crystal structure of TMPRSS2 in complex with Camostat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Duan, Y, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-06-04 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
