8J8W
 
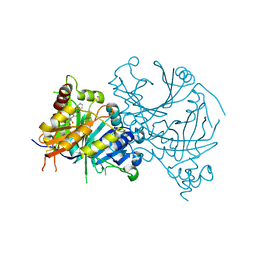 | | Crystal structure of AtHPPD-Y19788 complex | | Descriptor: | 2,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-phenyl-quinazolin-4-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2023-05-02 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal structure of AtHPPD-Y19788 complex
To Be Published
|
|
5IMX
 
 | |
3K8A
 
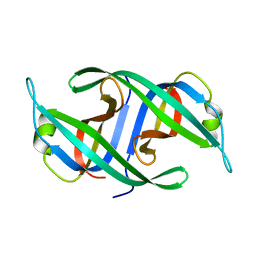 | | Neisseria gonorrhoeae PriB | | Descriptor: | Putative primosomal replication protein | | Authors: | Lopper, M.E, Dong, J, George, N.P, Duckett, K.L, DeBeer, M.A. | | Deposit date: | 2009-10-14 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of Neisseria gonorrhoeae PriB reveals mechanistic differences among bacterial DNA replication restart pathways
Nucleic Acids Res., 38, 2010
|
|
1XF7
 
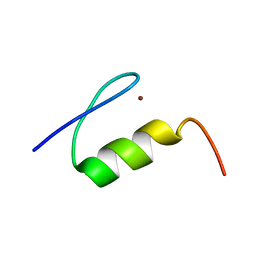 | | High Resolution NMR Structure of the Wilms' Tumor Suppressor Protein (WT1) Finger 3 | | Descriptor: | Wilms' Tumor Protein, ZINC ION | | Authors: | Lachenmann, M.J, Ladbury, J.E, Dong, J, Huang, K, Carey, P, Weiss, M.A. | | Deposit date: | 2004-09-14 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Why zinc fingers prefer zinc: ligand-field symmetry and the hidden thermodynamics of metal ion selectivity
Biochemistry, 43, 2004
|
|
5D1I
 
 | |
8K0B
 
 | | Cryo-EM structure of TMEM63C | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Qin, Y, Yu, D, Dong, J, Dang, S. | | Deposit date: | 2023-07-08 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM structure of TMEM63C suggests it functions as a monomer.
Nat Commun, 14, 2023
|
|
4M1K
 
 | |
4MYU
 
 | |
4MYT
 
 | |
3T4F
 
 | | Crystal Structure of a KGE Collagen Mimetic Peptide at 1.68 A | | Descriptor: | collagen mimetic peptide | | Authors: | Fallas, J.A, Dong, J, Miller, M.D, Tao, Y.J, Hartgerink, J.D. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural insights into charge pair interactions in triple helical collagen-like proteins.
J.Biol.Chem., 287, 2012
|
|
3U29
 
 | | Crystal Structure of a KGD Collagen Mimetic Peptide at 2.0 A | | Descriptor: | collagen mimetic peptide | | Authors: | Fallas, J.A, Dong, J, Miller, M.D, Tao, Y.J, Hartgerink, J.D. | | Deposit date: | 2011-10-02 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into charge pair interactions in triple helical collagen-like proteins.
J.Biol.Chem., 287, 2012
|
|
4OBY
 
 | | Crystal Structure of E.coli Arginyl-tRNA Synthetase and Ligand Binding Studies Revealed Key Residues in Arginine Recognition | | Descriptor: | ARGININE, Arginine--tRNA ligase | | Authors: | Bi, K, Zheng, Y, Dong, J, Gao, F, Wang, J, Wang, Y, Gong, W. | | Deposit date: | 2014-01-08 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Crystal structure of E. coli arginyl-tRNA synthetase and ligand binding studies revealed key residues in arginine recognition.
Protein Cell, 5, 2014
|
|
4Q2V
 
 | | Crystal Structure of Ricin A chain complexed with Baicalin inhibitor | | Descriptor: | 5,6-dihydroxy-4-oxo-2-phenyl-4H-chromen-7-yl beta-D-glucopyranosiduronic acid, Ricin | | Authors: | Deng, X, Li, X, Dong, J, Chen, Y. | | Deposit date: | 2014-04-10 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Baicalin inhibits the lethality of ricin in mice by inducing protein oligomerization.
J.Biol.Chem., 290, 2015
|
|
6PCK
 
 | | Crystal structure of human diphosphoinositol polyphosphate phosphohydrolase 1 in complex with 1-IP7 | | Descriptor: | (1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Dollins, D.E, Neubauer, J, Dong, J, York, J.D. | | Deposit date: | 2019-06-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Vip1 is a kinase and pyrophosphatase switch that regulates inositol diphosphate signaling.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PCL
 
 | | Crystal structure of human diphosphoinositol polyphosphate phosphohydrolase 1 in complex with 5-IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Dollins, D.E, Neubauer, J, Dong, J, York, J.D. | | Deposit date: | 2019-06-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Vip1 is a kinase and pyrophosphatase switch that regulates inositol diphosphate signaling.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7XVH
 
 | | Crystal structure of AtHPPD-Y13287 complex | | Descriptor: | 1,5-dimethyl-3-(2-methylphenyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-05-23 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
7X5U
 
 | | Crystal structure of AtHPPD-diketonitrile complex | | Descriptor: | (1R,2S,3R)-1-cyclopropyl-2-(iminomethyl)-3-[2-methylsulfonyl-4-(trifluoromethyl)phenyl]propane-1,3-diol, (4S)-2-METHYL-2,4-PENTANEDIOL, 4-hydroxyphenylpyruvate dioxygenase, ... | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-05 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
7X64
 
 | | Crystal structure of AtHPPD-Y18027 complex | | Descriptor: | 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-(phenylmethyl)quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-06 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
7X6N
 
 | | Crystal structure of AtHPPD-Y191710 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, ethyl 5-methyl-4-oxidanylidene-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-phenyl-quinazoline-2-carboxylate | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-07 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.691 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
7X7L
 
 | | Crystal structure of ZmHPPD-Y13161 complex | | Descriptor: | 3-(2,6-dimethylphenyl)-1-methyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-09 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.887 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
7X5W
 
 | | Crystal structure of AtHPPD-Y18022 complex | | Descriptor: | 1,5-dimethyl-3-naphthalen-1-yl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-05 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
7X5Y
 
 | | Crystal structure of AtHPPD-Y14240 complex | | Descriptor: | (1R,3S)-5-methyl-2-[(1S)-2-naphthalen-1-yloxy-1-oxidanyl-ethyl]cyclohexane-1,3-diol, (4S)-2-METHYL-2,4-PENTANEDIOL, 4-hydroxyphenylpyruvate dioxygenase, ... | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-05 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
7X69
 
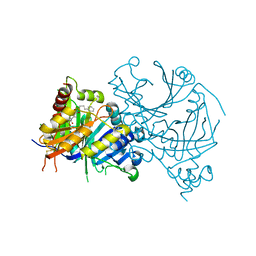 | | Crystal structure of AtHPPD-Y191053 complex | | Descriptor: | 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-(4-phenylbutyl)quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-06 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
7X5Z
 
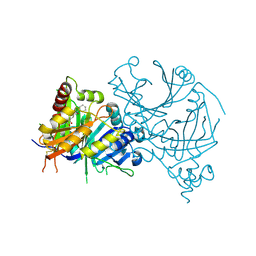 | | Crystal structure of AtHPPD-Y13162 complex | | Descriptor: | 1-methyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-phenyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-05 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.688 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
7X5S
 
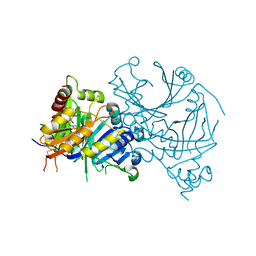 | | Crystal structure of AtHPPD-Y14157 complex | | Descriptor: | 2-methyl-4-(2-nitro-4-piperidin-1-yl-phenyl)carbonyl-5-phenyl-1H-pyrazol-3-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-05 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.706 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
