5XOB
 
 | | Crystal structure of apo TiaS (tRNAIle2 agmatidine synthetase) | | Descriptor: | MAGNESIUM ION, ZINC ION, tRNA(Ile2) 2-agmatinylcytidine synthetase TiaS | | Authors: | Dong, J. | | Deposit date: | 2017-05-27 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of tRNA-Modifying Enzyme TiaS and Motions of Its Substrate Binding Zinc Ribbon.
J. Mol. Biol., 430, 2018
|
|
4Y49
 
 | |
3QSQ
 
 | |
4GF6
 
 | | crystal structure of GFP with cuprum bound at the Incorporated metal Chelating Amino Acid PYZ151 | | Descriptor: | CALCIUM ION, COPPER (II) ION, green fluorescent protein | | Authors: | Dong, J, Liu, X, Li, J, Wang, J, Gong, W. | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Genetic incorporation of a metal-chelating amino Acid as a probe for protein electron transfer.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4GES
 
 | | crystal structure of GFP-TYR151PYZ with an unnatural amino acid incorporation | | Descriptor: | Green fluorescent protein | | Authors: | Dong, J, Liu, X, Li, J, Wang, J, Gong, W. | | Deposit date: | 2012-08-02 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Genetic incorporation of a metal-chelating amino Acid as a probe for protein electron transfer.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4TS8
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with XZ08 | | Descriptor: | 1,2-ETHANEDIOL, 4-(1-acetyl-1H-indol-3-yl)-5-methyl-1,2-dihydro-3H-pyrazol-3-one, CREB-binding protein | | Authors: | Dong, J, Gartenmann, L, Spiliotopoulos, D, Caflisch, A. | | Deposit date: | 2014-06-18 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with XZ08
To Be Published
|
|
4TWO
 
 | | Human EphA3 Kinase domain in complex with compound 164 | | Descriptor: | 5-{[3-carbamoyl-4-(3,4-dimethylphenyl)-5-methylthiophen-2-yl]amino}-5-oxopentanoic acid, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-07-01 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural Analysis of the Binding of Type I, I1/2, and II Inhibitors to Eph Tyrosine Kinases.
Acs Med.Chem.Lett., 6, 2015
|
|
4TWN
 
 | | Human EphA3 Kinase domain in complex with Birb796 | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-07-01 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.706 Å) | | Cite: | Structural Analysis of the Binding of Type I, I1/2, and II Inhibitors to Eph Tyrosine Kinases.
Acs Med.Chem.Lett., 6, 2015
|
|
4GK3
 
 | | Human EphA3 Kinase domain in complex with ligand 87 | | Descriptor: | 8-butyl-1-methyl-7-(2-methylphenyl)-1H-imidazo[2,1-f]purine-2,4(3H,8H)-dione, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2012-08-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Optimization of Inhibitors of the Tyrosine Kinase EphB4. 2. Cellular Potency Improvement and Binding Mode Validation by X-ray Crystallography.
J.Med.Chem., 56, 2013
|
|
4GK2
 
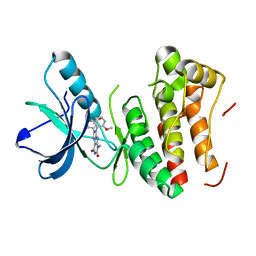 | | Human EphA3 Kinase domain in complex with ligand 66 | | Descriptor: | 7-(5-hydroxy-2-methylphenyl)-8-(2-methoxyphenyl)-1-methyl-1H-imidazo[2,1-f]purine-2,4(3H,8H)-dione, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2012-08-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Optimization of Inhibitors of the Tyrosine Kinase EphB4. 2. Cellular Potency Improvement and Binding Mode Validation by X-ray Crystallography.
J.Med.Chem., 56, 2013
|
|
4GK4
 
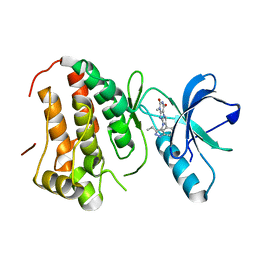 | | Human EphA3 Kinase domain in complex with ligand 90 | | Descriptor: | 8-butyl-1-methyl-7-(5-methyl-1H-indazol-4-yl)-1H-imidazo[2,1-f]purine-2,4(3H,8H)-dione, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2012-08-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimization of Inhibitors of the Tyrosine Kinase EphB4. 2. Cellular Potency Improvement and Binding Mode Validation by X-ray Crystallography.
J.Med.Chem., 56, 2013
|
|
4G2F
 
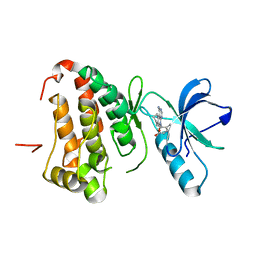 | | Human EphA3 kinase domain in complex with compound 7 | | Descriptor: | 1-amino-5-(5-hydroxy-2-methylphenyl)-7,8,9,10-tetrahydropyrimido[4,5-c]isoquinolin-6(5H)-one, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2012-07-12 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Discovery of a novel chemotype of tyrosine kinase inhibitors by fragment-based docking and molecular dynamics.
ACS MED.CHEM.LETT., 3, 2012
|
|
7L7E
 
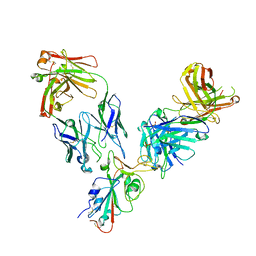 | |
7L7D
 
 | |
5OCB
 
 | | Crystal structure of nitric oxide bound D97N mutant of three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, HEME C, NITRIC OXIDE, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
5OCF
 
 | | Crystal structure of nitric oxide bound to three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, HEME C, NITRIC OXIDE, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
5OBO
 
 | | Crystal structure of nitrite bound D97N mutant of three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, GLYCEROL, HEME C, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-28 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
4XPD
 
 | |
4XNH
 
 | |
6UIG
 
 | |
5JRP
 
 | | crystal structure of monoclonal antibody MR78 Fab | | Descriptor: | SODIUM ION, marberg virus monoclonal antibody MR78 Fab heavy chain, marberg virus monoclonal antibody MR78 Fab light chain | | Authors: | Dong, J, Crowe, J. | | Deposit date: | 2016-05-06 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Non-local Interactions between CDR Loops in Binding Affinity of MR78 Antibody to Marburg Virus Glycoprotein.
Structure, 25, 2017
|
|
6VY4
 
 | |
6VY5
 
 | |
6VY6
 
 | |
6P9J
 
 | | crystal structure of human anti staphylococcus aureus antibody STAU-229 Fab | | Descriptor: | TRIS(HYDROXYETHYL)AMINOMETHANE, human anti staphylococcus aureus antibody STAU-229 Fab heavy chain, human anti staphylococcus aureus antibody STAU-229 Fab light chain | | Authors: | Dong, J, Crowe, J.E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human V H 1-69 Gene-Encoded Human Monoclonal Antibodies against Staphylococcus aureus IsdB Use at Least Three Distinct Modes of Binding To Inhibit Bacterial Growth and Pathogenesis.
Mbio, 10, 2019
|
|
