4M05
 
 | | Crystal Structure of Mutant Chlorite Dismutase from Candidatus Nitrospira defluvii R173E | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Chlorite dismutase, ... | | Authors: | Gysel, K, Hagmueller, A, Djinovic-Carugo, K. | | Deposit date: | 2013-08-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Manipulating conserved heme cavity residues of chlorite dismutase: effect on structure, redox chemistry, and reactivity.
Biochemistry, 53, 2014
|
|
7OWI
 
 | | Crystal structure of dimeric chlorite dismutase variant R127A (CCld R127A) from Cyanothece sp. PCC7425 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Chlorite dismutase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2021-06-18 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Impact of the dynamics of the catalytic arginine on nitrite and chlorite binding by dimeric chlorite dismutase.
J.Inorg.Biochem., 227, 2021
|
|
7OU9
 
 | | Crystal structure of dimeric chlorite dismutase variant Q74E (CCld Q74E) from Cyanothece sp. PCC7425 in complex with nitrite | | Descriptor: | Chlorite dismutase, GLYCEROL, NITRITE ION, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2021-06-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Impact of the dynamics of the catalytic arginine on nitrite and chlorite binding by dimeric chlorite dismutase.
J.Inorg.Biochem., 227, 2021
|
|
7OUA
 
 | | Crystal structure of dimeric chlorite dismutase variant R127K (CCld R127K) from Cyanothece sp. PCC7425 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chlorite dismutase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2021-06-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Impact of the dynamics of the catalytic arginine on nitrite and chlorite binding by dimeric chlorite dismutase.
J.Inorg.Biochem., 227, 2021
|
|
7OUY
 
 | | Crystal structure of dimeric chlorite dismutase variant R127A (CCld R127A) from Cyanothece sp. PCC7425 in complex with nitrite | | Descriptor: | Chlorite dismutase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2021-06-14 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Impact of the dynamics of the catalytic arginine on nitrite and chlorite binding by dimeric chlorite dismutase.
J.Inorg.Biochem., 227, 2021
|
|
5O4E
 
 | | Crystal structure of VEGF in complex with heterodimeric Fcab JanusCT6 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Mlynek, G, Lobner, E, Kubinger, K, Humm, A, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Two-faced Fcab prevents polymerization with VEGF and reveals thermodynamics and the 2.15 angstrom crystal structure of the complex.
MAbs, 9, 2017
|
|
5LOQ
 
 | | Structure of coproheme bound HemQ from Listeria monocytogenes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Putative heme-dependent peroxidase lmo2113, ... | | Authors: | Puehringer, D, Mlynek, G, Hofbauer, S, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Hydrogen peroxide-mediated conversion of coproheme to heme b by HemQ-lessons from the first crystal structure and kinetic studies.
FEBS J., 283, 2016
|
|
5NL6
 
 | |
5NKV
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 at pH 9.0 and 293 K. | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2017-04-03 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5NL7
 
 | | The crystal structure of the Actin Binding Domain (ABD) of alpha actinin from Entamoeba histolytica | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Calponin homology domain protein putative | | Authors: | Pinotsis, N, Djinovic-Carugo, K, Khan, M.B. | | Deposit date: | 2017-04-04 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Calcium modulates the domain flexibility and function of an alpha-actinin similar to the ancestral alpha-actinin.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5MFA
 
 | | Crystal structure of human promyeloperoxidase (proMPO) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grishkovskaya, I, Furtmueller, P.G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-11-17 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of human promyeloperoxidase (proMPO) and the role of the propeptide in processing and maturation.
J. Biol. Chem., 292, 2017
|
|
7OU5
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 in complex with nitrite | | Descriptor: | Chlorite Dismutase, GLYCEROL, NITRITE ION, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2021-06-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Impact of the dynamics of the catalytic arginine on nitrite and chlorite binding by dimeric chlorite dismutase.
J.Inorg.Biochem., 227, 2021
|
|
7OU7
 
 | | Crystal structure of dimeric chlorite dismutase variant Q74V (CCld Q74V) from Cyanothece sp. PCC7425 in complex with nitrite | | Descriptor: | Chlorite dismutase, GLYCEROL, NITRITE ION, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2021-06-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Impact of the dynamics of the catalytic arginine on nitrite and chlorite binding by dimeric chlorite dismutase.
J.Inorg.Biochem., 227, 2021
|
|
7OUV
 
 | |
7OUU
 
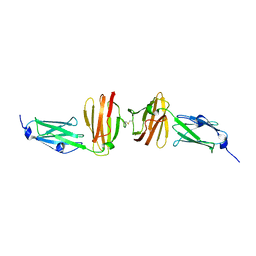 | |
7P0E
 
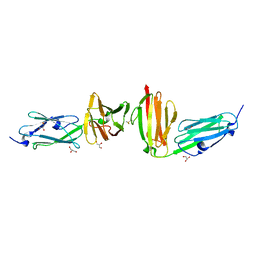 | |
4JG3
 
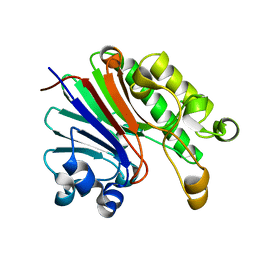 | | Crystal structure of catabolite repression control protein (crc) from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, Catabolite repression control protein | | Authors: | Grishkovskaya, I, Milojevic, T, Sonnleitner, E, Blaesi, U, Djinovic-Carugo, K. | | Deposit date: | 2013-02-28 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Pseudomonas aeruginosa Catabolite Repression Control Protein Crc Is Devoid of RNA Binding Activity
Plos One, 8, 2013
|
|
5MAU
 
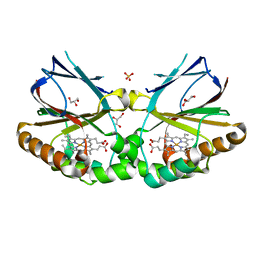 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 (pH 6.5) | | Descriptor: | Chlorite dismutase, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-11-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5NKU
 
 | | Joint neutron/X-ray structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2017-04-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5JXF
 
 | | Crystal structure of Flavobacterium psychrophilum DPP11 in complex with dipeptide Arg-Asp | | Descriptor: | ARGININE, ASPARTIC ACID, Asp/Glu-specific dipeptidyl-peptidase, ... | | Authors: | Bezerra, G.A, Fedosyuk, S, Ohara-Nemoto, Y, Nemoto, T.K, Djinovic-Carugo, K. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5JWG
 
 | | Crystal structure of Porphyromonas endodontalis DPP11 in complex with dipeptide Arg-Asp | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, ASPARTIC ACID, ... | | Authors: | Bezerra, G.A, Fedosyuk, S, Ohara-Nemoto, Y, Nemoto, T.K, Djinovic-Carugo, K. | | Deposit date: | 2016-05-12 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5K65
 
 | | Crystal structure of VEGF binding IgG1-Fc (Fcab CT6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Humm, A, Lobner, E, Kitzmuller, M, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two-faced Fcab prevents polymerization with VEGF and reveals thermodynamics and the 2.15 angstrom crystal structure of the complex.
MAbs, 9, 2017
|
|
6RWV
 
 | | Structure of apo-LmCpfC | | Descriptor: | Ferrochelatase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Hofbauer, S, Helm, J, Djinovic-Carugo, K, Furtmueller, P.G. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6386379 Å) | | Cite: | Crystal structures and calorimetry reveal catalytically relevant binding mode of coproporphyrin and coproheme in coproporphyrin ferrochelatase.
Febs J., 287, 2020
|
|
5JY0
 
 | | Crystal structure of Porphyromonas endodontalis DPP11 in complex with substrate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Asp/Glu-specific dipeptidyl-peptidase, LEU-ASP-VAL | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5JWF
 
 | | Crystal structure of Porphyromonas gingivalis DPP11 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, GLYCEROL, S-1,2-PROPANEDIOL | | Authors: | Bezerra, G.A, Fedosyuk, S, Ohara-Nemoto, Y, Nemoto, T.K, Djinovic-Carugo, K. | | Deposit date: | 2016-05-12 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
