7XT2
 
 | | Crystal structure of TRIM72 | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Zhou, C, Ma, Y.M, Ding, L. | | Deposit date: | 2022-05-15 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for TRIM72 oligomerization during membrane damage repair.
Nat Commun, 14, 2023
|
|
3MJ0
 
 | |
5ZBZ
 
 | | Crystal structure of the DEAD domain of Human eIF4A with sanguinarine | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, Eukaryotic initiation factor 4A-I, MALONATE ION | | Authors: | Ding, Y, Ding, L. | | Deposit date: | 2018-02-14 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.30860257 Å) | | Cite: | Targeting the N Terminus of eIF4AI for Inhibition of Its Catalytic Recycling.
Cell Chem Biol, 26, 2019
|
|
6ITQ
 
 | | Crystal structure of cortisol complexed with its nanobody at pH 10.5 | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, SULFATE ION, anti-cortisol camelid antibody | | Authors: | Ding, Y, Ding, L.L, Wang, Z.Y, Zhong, P.Y. | | Deposit date: | 2018-11-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.526 Å) | | Cite: | Structural insights into the mechanism of single domain VHH antibody binding to cortisol.
Febs Lett., 593, 2019
|
|
6ITP
 
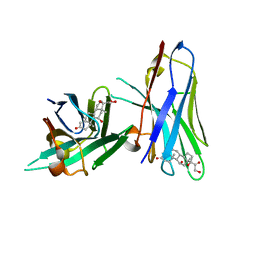 | | Crystal structure of cortisol complexed with its nanobody at pH 3.5 | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, anti-cortisol camelid antibody | | Authors: | Ding, Y, Ding, L.L, Wang, Z.Y, Zhong, P.Y. | | Deposit date: | 2018-11-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Structural insights into the mechanism of single domain VHH antibody binding to cortisol.
Febs Lett., 593, 2019
|
|
3QKW
 
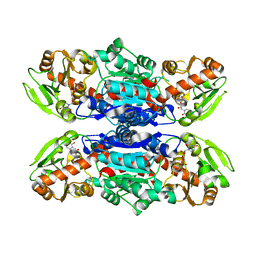 | | Structure of Streptococcus parasangunini Gtf3 glycosyltransferase | | Descriptor: | Nucleotide sugar synthetase-like protein, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhu, F, Erlandsen, H, Huang, Y, Ding, L, Zhou, M, Liang, X, Ma, J.-B, Wu, H. | | Deposit date: | 2011-02-01 | | Release date: | 2011-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Structural and Functional Analysis of a New Subfamily of Glycosyltransferases Required for Glycosylation of Serine-rich Streptococcal Adhesins.
J.Biol.Chem., 286, 2011
|
|
6XBC
 
 | |
6XBB
 
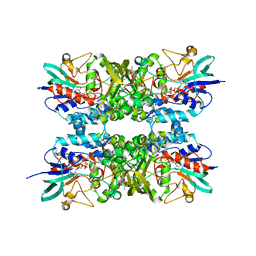 | |
7TXG
 
 | | Structure of the Class II Fructose-1,6-Bisphosphatase from Francisella tularensis with native Mn++ divalent cation and partially occupied product F6P | | Descriptor: | Fructose-1,6-bisphosphatase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Abad-Zapatero, C, Selezneva, A.I, Harding, L.N.M, Movahedzadeh, F. | | Deposit date: | 2022-02-09 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New structures of Class II Fructose-1,6-Bisphosphatase from Francisella tularensis provide a framework for a novel catalytic mechanism for the entire class.
Plos One, 18, 2023
|
|
7SWL
 
 | | CryoEM structure of the N-terminal-deleted Rix7 AAA-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kocaman, S, Stanley, R.E, Lo, Y.H, Krahn, J, Dandey, V.P, Sobhany, M, Petrovich, M, Williams, J.G, Deterding, L.J, Borgnia, M.J, Etigunta, S. | | Deposit date: | 2021-11-20 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
7T0V
 
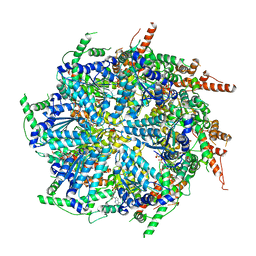 | | CryoEM structure of the crosslinked Rix7 AAA-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kocaman, S, Stanley, R.E, Lo, Y.H, Krahn, J, Dandey, V.P, Sobhany, M, Petrovich, M, Williams, J.G, Deterding, L.J, Borgnia, M.J, Etigunta, S. | | Deposit date: | 2021-11-30 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
4FA0
 
 | | Crystal structure of human AdPLA to 2.65 A resolution | | Descriptor: | Group XVI phospholipase A1/A2 | | Authors: | Lovell, S, Battaile, K.P, Addington, L, Zhang, N, Rao, J.L.U.M, Moise, A.R. | | Deposit date: | 2012-05-21 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure/Function relationships of adipose phospholipase A2 containing a cys-his-his catalytic triad.
J.Biol.Chem., 287, 2012
|
|
1DLB
 
 | | HELICAL INTERACTIONS IN THE HIV-1 GP41 CORE REVEALS STRUCTURAL BASIS FOR THE INHIBITORY ACTIVITY OF GP41 PEPTIDES | | Descriptor: | HIV-1 ENVELOPE GLYCOPROTEIN GP41 | | Authors: | Shu, W, Liu, J, Ji, H, Rading, L, Jiang, S, Lu, M. | | Deposit date: | 1999-12-09 | | Release date: | 1999-12-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Helical interactions in the HIV-1 gp41 core reveal structural basis for the inhibitory activity of gp41 peptides.
Biochemistry, 39, 2000
|
|
2VAQ
 
 | | STRUCTURE OF STRICTOSIDINE SYNTHASE IN COMPLEX WITH INHIBITOR | | Descriptor: | (2S,3R,4S)-methyl 4-(2-(2-(1H-indol-3-yl)ethylamino)ethyl)-2-((2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yloxy)-3-vinyl-3,4-dihydro-2H-pyran-5-carboxylate, STRICTOSIDINE SYNTHASE | | Authors: | Maresh, J, Giddings, L.A, Friedrich, A, Loris, E.A, Panjikar, S, Trout, B.L, Stoeckigt, J, Peters, B, O'Connor, S.E. | | Deposit date: | 2007-09-04 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Strictosidine synthase: mechanism of a Pictet-Spengler catalyzing enzyme.
J. Am. Chem. Soc., 130, 2008
|
|
