6BU7
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor RD130 1-[2-(Piperidin-4-yl)ethyl]-5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole | | Descriptor: | 1-[2-(piperidin-4-yl)ethyl]-5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bryson, S, De Gasparo, R, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2017-12-08 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Biological Evaluation and X-ray Co-crystal Structures of Cyclohexylpyrrolidine Ligands for Trypanothione Reductase, an Enzyme from the Redox Metabolism of Trypanosoma.
ChemMedChem, 13, 2018
|
|
6BTL
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor RD117 1-[2-(Piperazin-1-yl)ethyl]-5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole | | Descriptor: | 1-[2-(piperazin-1-yl)ethyl]-5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bryson, S, De Gasparo, R, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2017-12-06 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Biological Evaluation and X-ray Co-crystal Structures of Cyclohexylpyrrolidine Ligands for Trypanothione Reductase, an Enzyme from the Redox Metabolism of Trypanosoma.
ChemMedChem, 13, 2018
|
|
6GXT
 
 | | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography: FAcD2052MS after reaction initiation | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Mueller-Werkmeister, H, Tellkamp, F, Stuart, W, Persch, E, De Gasparo, R, Diederich, F, Pai, E.F, Miller, R.J.D. | | Deposit date: | 2018-06-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography.
Nat. Methods, 15, 2018
|
|
6GXD
 
 | | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography: FAcD752MS after reaction initiation | | Descriptor: | CHLORIDE ION, fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Mueller-Werkmeister, H, Tellkamp, F, Stuart, W, Persch, E, De Gasparo, R, Diederich, F, Pai, E.F, Miller, R.J.D. | | Deposit date: | 2018-06-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography.
Nat. Methods, 15, 2018
|
|
6GXH
 
 | | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography: FAcD 0MS after reaction initiation | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Schulz, E.C, Mehrabi, P, Mueller-Werkmeister, H, Tellkamp, F, Stuart, W, Persch, E, De Gasparo, R, Diederich, F, Pai, E.F, Miller, R.J.D. | | Deposit date: | 2018-06-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography.
Nat. Methods, 15, 2018
|
|
6GXL
 
 | | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography: RADDAM2 | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Schulz, E.C, Mehrabi, P, Mueller-Werkmeister, H, Tellkamp, F, Stuart, W, Persch, E, De Gasparo, R, Diederich, F, Pai, E.F, Miller, R.J.D. | | Deposit date: | 2018-06-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography.
Nat. Methods, 15, 2018
|
|
6GXF
 
 | | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography: RADDAM1 | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Schulz, E.C, Mehrabi, P, Mueller-Werkmeister, H, Tellkamp, F, Stuart, W, Persch, E, De Gasparo, R, Diederich, F, Pai, E.F, Miller, R.J.D. | | Deposit date: | 2018-06-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography.
Nat. Methods, 15, 2018
|
|
2QZR
 
 | | tRNA-Guanine Transglycosylase(TGT) in Complex with 6-amino-2-[(1-naphthylmethyl)amino]-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-2-[(1-naphthylmethyl)amino]-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Hoertner, S.R, Ritschel, T, Stengl, B, Kramer, C, Schweizer, W.B, Wagner, B, Kansy, M, Klebe, G, Diederich, F. | | Deposit date: | 2007-08-17 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Potent Inhibitors of tRNA-Guanine Transglycosylase, an Enzyme Linked to the Pathogenicity of the Shigella Bacterium: Charge-Assisted Hydrogen Bonding.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
2GZL
 
 | | Structure of 2C-methyl-D-erythritol 2,4-clycodiphosphate synthase complexed with a CDP derived fluorescent inhibitor | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 5'-O-{[({[2-({[5-(DIMETHYLAMINO)NAPHTHALEN-1-YL]SULFONYL}AMINO)ETHYL]OXY}PHOSPHINATO)OXY]PHOSPHINATO}CYT, GERANYL DIPHOSPHATE, ... | | Authors: | Crane, C.M, Kaiser, J, Ramsden, N.L, Lauw, S, Rohdich, F, Wolfgang, E, Hunter, W.N, Bacher, A, Diederich, F. | | Deposit date: | 2006-05-11 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fluorescent Inhibitors for Ispf, an Enzyme in the Non-Mevalonate Pathway for Isoprenoid Biosynthesis and a Potential Target for Antimalarial Therapy.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
6OEX
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor 3-(2-{1-[2-(Piperidin-4-yl)ethyl]-1H-indol-5-yl}-5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3- thiazol-4-yl)-N-(2,2,2-trifluoroethyl)prop-2-yn-1-amine | | Descriptor: | 3-(2-{1-[2-(piperidin-4-yl)ethyl]-1H-indol-5-yl}-5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-4-yl)-N-(2,2,2-trifluoroethyl)prop-2-yn-1-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Halgas, O, De Gasparo, R, Harangozo, D, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting a Large Active Site: Structure-Based Design of Nanomolar Inhibitors of Trypanosoma brucei Trypanothione Reductase.
Chemistry, 25, 2019
|
|
6OEY
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor (+)-5-{5-[1-(Pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1-{[(2S)-pyrrolidin-2-yl]methyl}-1H-indole | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1-{[(2S)-pyrrolidin-2-yl]methyl}-1H-indole, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Halgas, O, De Gasparo, R, Harangozo, D, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting a Large Active Site: Structure-Based Design of Nanomolar Inhibitors of Trypanosoma brucei Trypanothione Reductase.
Chemistry, 25, 2019
|
|
6OEZ
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor (+)-N-(Cyclobutylmethyl)-3-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-2-(1-{[(2S)-pyrro-lidin-2-yl]methyl}-1H-indol-5-yl)-1,3-thiazol-4-yl}prop-2-yn-1-amine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, N-(cyclobutylmethyl)-3-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-2-(1-{[(2S)-pyrrolidin-2-yl]methyl}-1H-indol-5-yl)-1,3-thiazol-4-yl}prop-2-yn-1-amine, ... | | Authors: | Halgas, O, De Gasparo, R, Harangozo, D, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting a Large Active Site: Structure-Based Design of Nanomolar Inhibitors of Trypanosoma brucei Trypanothione Reductase.
Chemistry, 25, 2019
|
|
1K4G
 
 | | CRYSTAL STRUCTURE OF TRNA-GUANINE TRANSGLYCOSYLASE (TGT) COMPLEXED WITH 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE | | Descriptor: | 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE, TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Brenk, R, Meyer, E.A, Castellano, R.K, Furler, M, Stubbs, M.T, Klebe, G, Diederich, F. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo design, synthesis, and in vitro evaluation of inhibitors for prokaryotic tRNA-guanine transglycosylase: a dramatic sulfur effect on binding affinity.
ChemBioChem, 3, 2002
|
|
1K4H
 
 | | CRYSTAL STRUCTURE OF TRNA-GUANINE TRANSGLYCOSYLASE (TGT) COMPLEXED WITH 2,6-Diamino-8-propylsulfanylmethyl-3H-quinazoline-4-one | | Descriptor: | 2,6-DIAMINO-8-PROPYLSULFANYLMETHYL-3H-QUINAZOLINE-4-ONE, TRNA-GUANINE-TRANSGLYCOSYLASE, ZINC ION | | Authors: | Brenk, R, Meyer, E.A, Castellano, R.K, Furler, M, Stubbs, M.T, Klebe, G, Diederich, F. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design, synthesis, and in vitro evaluation of inhibitors for prokaryotic tRNA-guanine transglycosylase: a dramatic sulfur effect on binding affinity.
Chembiochem, 3, 2002
|
|
2JKH
 
 | | Factor Xa - cation inhibitor complex | | Descriptor: | 3-[(3~{a}~{S},4~{R},8~{a}~{S},8~{b}~{R})-4-[5-(5-chloranylthiophen-2-yl)-1,2-oxazol-3-yl]-1,3-bis(oxidanylidene)-4,6,7,8,8~{a},8~{b}-hexahydro-3~{a}~{H}-pyrrolo[3,4-a]pyrrolizin-2-yl]propyl-trimethyl-azanium, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Salonen, L.M, Bucher, C, Banner, D.W, Benz, J, Haap, W, Mary, J.L, Schweizer, W.B, Seiler, P, Kuster, O, Diederich, F. | | Deposit date: | 2008-08-28 | | Release date: | 2009-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Cation-Pi Interactions at the Active Site of Factor Xa: Dramatic Enhancement Upon Stepwise N-Alkylation of Ammonium Ions.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2BBF
 
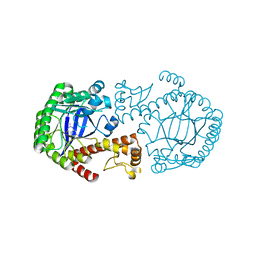 | | Crystal structure of tRNA-guanine transglycosylase (TGT) from Zymomonas mobilis in complex with 6-amino-3,7-dihydro-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-AMINO-3,7-DIHYDRO-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, ZINC ION, tRNA guanine transglycosylase | | Authors: | Stengl, B, Meyer, E.A, Heine, A, Brenk, R, Diederich, F, Klebe, G. | | Deposit date: | 2005-10-17 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding.
J.Mol.Biol., 370, 2007
|
|
1S38
 
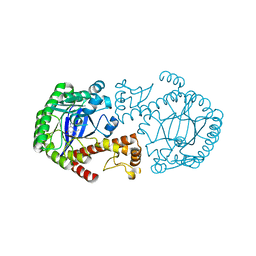 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2-AMINO-8-METHYLQUINAZOLIN-4(3H)-ONE | | Descriptor: | 2-AMINO-8-METHYLQUINAZOLIN-4(3H)-ONE, ZINC ION, tRNA guanine transglycosylase | | Authors: | Meyer, E.A, Furler, M, Diederich, F, Brenk, R, Klebe, G. | | Deposit date: | 2004-01-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Synthesis and In vitro Evaluation of 2-Aminoquinazolin-4(3H)-one-based Inhibitors for tRNA-Guanine Transglycosylase (TGT)
HELV.CHIM.ACTA, 87, 2004
|
|
4AXM
 
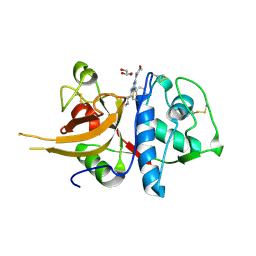 | | TRIAZINE CATHEPSIN INHIBITOR COMPLEX | | Descriptor: | 4-[(4-chlorobenzyl)(cyclohexyl)amino]-6-morpholin-4-yl-1,3,5-triazine-2-carboxamide, CATHEPSIN L1, GLYCEROL | | Authors: | Ehmke, V, Diederich, F, Banner, D.W, Benz, J. | | Deposit date: | 2012-06-13 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Optimization of Triazine Nitriles as Rhodesain Inhibitors: Structure-Activity Relationships, Bioisosteric Imidazopyridine Nitriles, and X-Ray Crystal Structure Analysis with Human Cathepsin L
Chemmedchem, 8, 2013
|
|
1Y5V
 
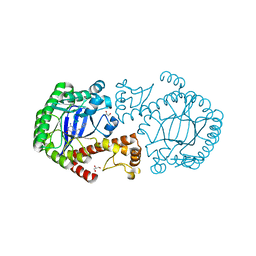 | | tRNA-Guanine Transglycosylase (TGT) in complex with 6-Amino-4-(2-phenylethyl)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-AMINO-4-(2-PHENYLETHYL)-1,7-DIHYDRO-8H-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Stengl, B, Meyer, E.A, Heine, A, Brenk, R, Diederich, F, Klebe, G. | | Deposit date: | 2004-12-03 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding
J.Mol.Biol., 370, 2007
|
|
6FSX
 
 | | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Schulz, E.C, Mehrabi, P, Mueller-werkmeiser, H, Tellkamp, F, Jha, A, Stuart, W, Persch, E, De Gasparo, R, Diederich, F, Pai, E, Miller, D. | | Deposit date: | 2018-02-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography.
Nat. Methods, 15, 2018
|
|
1Y5W
 
 | | tRNA-guanine Transglycosylase (TGT) in complex with 6-Amino-4-[2-(4-methylphenyl)ethyl]-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-AMINO-4-[2-(4-METHYLPHENYL)ETHYL]-1,7-DIHYDRO-8H-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Stengl, B, Meyer, E.A, Heine, A, Brenk, R, Diederich, F, Klebe, G. | | Deposit date: | 2004-12-03 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding
J.Mol.Biol., 370, 2007
|
|
1Y5X
 
 | | tRNA-guanine Transglycosylase (TGT) in complex with 6-Amino-4-[2-(4-methoxyphenyl)ethyl]-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-AMINO-4-[2-(4-METHOXYPHENYL)ETHYL]-1,7-DIHYDRO-8H-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Stengl, B, Meyer, E.A, Heine, A, Brenk, R, Diederich, F, Klebe, G. | | Deposit date: | 2004-12-03 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding
J.Mol.Biol., 370, 2007
|
|
5MRN
 
 | | Arabidopsis thaliana IspD Glu258Ala Mutant | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, CADMIUM ION, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
5XMR
 
 | | Plasmodium vivax SHMT(C346A) bound with PLP-glycine and GS395 | | Descriptor: | (4~{S})-6-azanyl-3-methyl-4-[3-[4-(phenylmethyl)sulfonylphenyl]-5-(trifluoromethyl)phenyl]-4-propan-2-yl-2~{H}-pyrano[2 ,3-c]pyrazole-5-carbonitrile, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-05-16 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Conformational Aspects in the Design of Inhibitors for Serine Hydroxymethyltransferase (SHMT): Biphenyl, Aryl Sulfonamide, and Aryl Sulfone Motifs
Chemistry, 23, 2017
|
|
5MRQ
 
 | | Arabidopsis thaliana IspD Asp262Ala Mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
