2AEB
 
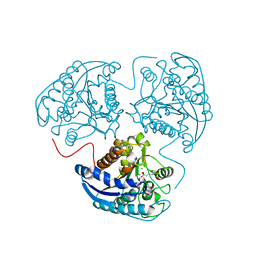 | | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in immune response. | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase 1, MANGANESE (II) ION | | Authors: | Di Costanzo, L, Sabio, G, Mora, A, Rodriguez, P.C, Ochoa, A.C, Centeno, F, Christianson, D.W. | | Deposit date: | 2005-07-21 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in the immune response.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1OVU
 
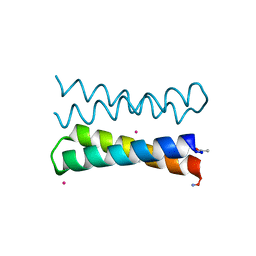 | |
3BV7
 
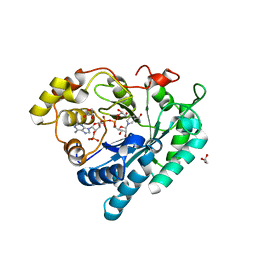 | | Crystal structure of Delta(4)-3-ketosteroid 5-beta-reductase in complex with NADP and glycerol. Resolution: 1.79 A. | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Di Costanzo, L, Drury, J, Penning, T.M, Christianson, D.W. | | Deposit date: | 2008-01-04 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of Human Liver {Delta}4-3-Ketosteroid 5{beta}-Reductase (AKR1D1) and Implications for Substrate Binding and Catalysis.
J.Biol.Chem., 283, 2008
|
|
3BUV
 
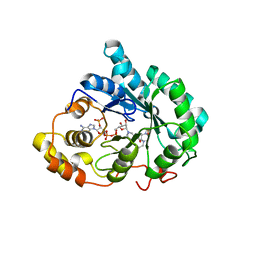 | | Crystal structure of human Delta(4)-3-ketosteroid 5-beta-reductase in complex with NADP and HEPES. Resolution: 1.35 A. | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Di Costanzo, L, Drury, J, Penning, T.M, Christianson, D.W. | | Deposit date: | 2008-01-03 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Human Liver {Delta}4-3-Ketosteroid 5{beta}-Reductase (AKR1D1) and Implications for Substrate Binding and Catalysis.
J.Biol.Chem., 283, 2008
|
|
1LT1
 
 | |
1OVV
 
 | |
1OVR
 
 | | CRYSTAL STRUCTURE OF FOUR-HELIX BUNDLE MODEL di-Mn(II)-DF1-L13 | | Descriptor: | MANGANESE (II) ION, four-helix bundle model di-Mn(II)-DF1-L13 | | Authors: | Di Costanzo, L, Geremia, S. | | Deposit date: | 2003-03-27 | | Release date: | 2004-05-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Response of a designed metalloprotein to changes in metal ion coordination, exogenous ligands, and active site volume determined by X-ray crystallography.
J.Am.Chem.Soc., 127, 2005
|
|
3SJT
 
 | | Crystal structure of human arginase I in complex with the inhibitor Me-ABH, Resolution 1.60 A, twinned structure | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5S)-5-amino-5-carboxyhexyl](trihydroxy)borate | | Authors: | Di Costanzo, L, Christianson, D.W. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Binding of alpha , alpha-disubstituted amino acids to arginase suggests new avenues for inhibitor design.
J.Med.Chem., 54, 2011
|
|
5SBG
 
 | |
1WVB
 
 | | Crystal structure of human arginase I: the mutant E256Q | | Descriptor: | Arginase 1, MANGANESE (II) ION, S-2-(BORONOETHYL)-L-CYSTEINE | | Authors: | Di Costanzo, L, Guadalupe, S, Mora, A, Centeno, F, Christianson, D.W. | | Deposit date: | 2004-12-14 | | Release date: | 2005-09-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human arginase I: the mutant E256Q
To be Published
|
|
1JM0
 
 | | CRYSTAL STRUCTURE OF FOUR-HELIX BUNDLE MODEL | | Descriptor: | DIMETHYL SULFOXIDE, MANGANESE (II) ION, PROTEIN (FOUR-HELIX BUNDLE MODEL) | | Authors: | Di Costanzo, L, Geremia, S. | | Deposit date: | 2001-07-17 | | Release date: | 2002-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Toward the de novo design of a catalytically active helix bundle: a substrate-accessible carboxylate-bridged dinuclear metal center.
J.Am.Chem.Soc., 123, 2001
|
|
1JMB
 
 | | CRYSTAL STRUCTURE OF FOUR-HELIX BUNDLE MODEL | | Descriptor: | DIMETHYL SULFOXIDE, MANGANESE (II) ION, PROTEIN (FOUR-HELIX BUNDLE MODEL) | | Authors: | Di Costanzo, L, Geremia, S. | | Deposit date: | 2001-07-18 | | Release date: | 2002-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Toward the de novo design of a catalytically active helix bundle: a substrate-accessible carboxylate-bridged dinuclear metal center.
J.Am.Chem.Soc., 123, 2001
|
|
1WVA
 
 | | Crystal structure of human arginase I from twinned crystal | | Descriptor: | Arginase 1, MANGANESE (II) ION, S-2-(BORONOETHYL)-L-CYSTEINE | | Authors: | Di Costanzo, L, Sabio, G, Mora, A, Rodriguez, P.C, Ochoa, A.C, Centeno, F, Christianson, D.W. | | Deposit date: | 2004-12-14 | | Release date: | 2005-09-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in the immune response
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5SBI
 
 | |
5SBJ
 
 | |
2Z6M
 
 | |
2ZAV
 
 | |
2PHA
 
 | |
2PLL
 
 | | Crystal structure of perdeuterated human arginase I | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, MANGANESE (II) ION, arginase-1 | | Authors: | Di Costanzo, L, Moulin, M, Haertlein, M, Meilleur, F, Christianson, D.W. | | Deposit date: | 2007-04-19 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expression, purification, assay, and crystal structure of perdeuterated human arginase I
Arch.Biochem.Biophys., 465, 2007
|
|
2PHO
 
 | |
3GMZ
 
 | |
2HG2
 
 | |
2IMP
 
 | | Crystal structure of lactaldehyde dehydrogenase from E. coli: the ternary complex with lactate (occupancy 0.5) and NADH. Crystals soaked with (L)-Lactate. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, LACTIC ACID, Lactaldehyde dehydrogenase, ... | | Authors: | Di Costanzo, L, Gomez, G.A, Christianson, D.W. | | Deposit date: | 2006-10-04 | | Release date: | 2007-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Lactaldehyde Dehydrogenase from Escherichia coli and Inferences Regarding Substrate and Cofactor Specificity.
J.Mol.Biol., 366, 2007
|
|
2ILU
 
 | |
1PYZ
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF MIMOCHROME IV | | Descriptor: | CHLORIDE ION, CO(III)-(DEUTEROPORPHYRIN IX), MIMOCHROME IV, ... | | Authors: | Di Costanzo, L, Geremia, S, Randaccio, L, Nastri, F, Maglio, O, Lombardi, A, Pavone, V. | | Deposit date: | 2003-07-09 | | Release date: | 2004-12-14 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Miniaturized heme proteins: crystal structure of Co(III)-mimochrome IV.
J.Biol.Inorg.Chem., 9, 2004
|
|
