2L04
 
 | | The Solution Structure of the C-terminal Ig-like Domain of the Bacteriophage Lambda Tail Tube Protein | | Descriptor: | Major tail protein V | | Authors: | Pell, L.G, Gasmi-Seabrook, G.M.C, Donaldson, L.W, Howell, P, Davidson, A.R, Maxwell, K.L. | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the C-Terminal Ig-like Domain of the Bacteriophage l Tail Tube Protein.
J.Mol.Biol., 403, 2010
|
|
2LCS
 
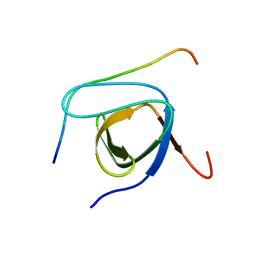 | | Yeast Nbp2p SH3 domain in complex with a peptide from Ste20p | | Descriptor: | NAP1-binding protein 2, Serine/threonine-protein kinase STE20 | | Authors: | Gorelik, M, Davidson, A.R. | | Deposit date: | 2011-05-07 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Distinct Peptide Binding Specificities of Src Homology 3 (SH3) Protein Domains Can Be Determined by Modulation of Local Energetics across the Binding Interface.
J.Biol.Chem., 287, 2012
|
|
2LTF
 
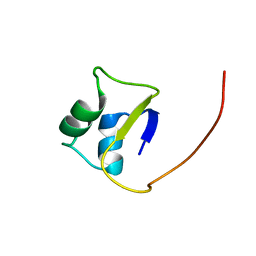 | |
6ARZ
 
 | | Structure of a phage anti-CRISPR protein | | Descriptor: | BROMIDE ION, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Calmettes, C, Shah, M, Pawluk, A, Davidson, A.R, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Disabling a Type I-E CRISPR-Cas Nuclease with a Bacteriophage-Encoded Anti-CRISPR Protein.
MBio, 8, 2017
|
|
3FZ2
 
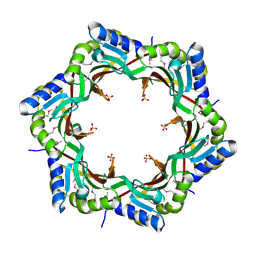 | | Crystal structure of the tail terminator protein from phage lambda (gpU-D74A) | | Descriptor: | Minor tail protein U, SULFATE ION | | Authors: | Pell, L.G, Liu, A, Edmonds, E, Donaldson, L.W, Howell, P.L, Davidson, A.R. | | Deposit date: | 2009-01-23 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages.
J.Mol.Biol., 389, 2009
|
|
3FZB
 
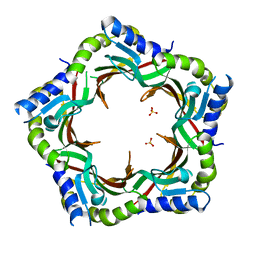 | | Crystal structure of the tail terminator protein from phage lambda (gpU-WT) | | Descriptor: | Minor tail protein U, SULFATE ION | | Authors: | Pell, L.G, Liu, A, Edmonds, E, Donaldson, L.W, Howell, P.L, Davidson, A.R. | | Deposit date: | 2009-01-24 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages.
J.Mol.Biol., 389, 2009
|
|
6AS4
 
 | | Structure of a phage anti-CRISPR protein | | Descriptor: | NHis AcrE1 anti-crispr protein | | Authors: | Shah, M, Calmettes, C, Pawluk, A, Mejdani, M, Davidson, A.R, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2017-08-23 | | Release date: | 2017-12-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disabling a Type I-E CRISPR-Cas Nuclease with a Bacteriophage-Encoded Anti-CRISPR Protein.
MBio, 8, 2017
|
|
6AS3
 
 | | Structure of a phage anti-CRISPR protein | | Descriptor: | NHis AcrE1 protein | | Authors: | Shah, M, Calmettes, C, Pawluk, A, Mejdani, M, Davidson, A.R, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disabling a Type I-E CRISPR-Cas Nuclease with a Bacteriophage-Encoded Anti-CRISPR Protein.
MBio, 8, 2017
|
|
7KIX
 
 | |
1HYW
 
 | | SOLUTION STRUCTURE OF BACTERIOPHAGE LAMBDA GPW | | Descriptor: | HEAD-TO-TAIL JOINING PROTEIN W | | Authors: | Maxwell, K.L, Yee, A.A, Booth, V, Arrowsmith, C.H, Gold, M, Davidson, A.R. | | Deposit date: | 2001-01-22 | | Release date: | 2001-04-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of bacteriophage lambda protein W, a small morphogenetic protein possessing a novel fold.
J.Mol.Biol., 308, 2001
|
|
1K0H
 
 | | Solution structure of bacteriophage lambda gpFII | | Descriptor: | gpFII | | Authors: | Maxwell, K.L, Yee, A.A, Arrowsmith, C.H, Gold, M, Davidson, A.R. | | Deposit date: | 2001-09-19 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the bacteriophage lambda head-tail joining protein, gpFII.
J.Mol.Biol., 318, 2002
|
|
5UZ9
 
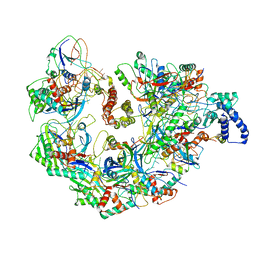 | | Cryo EM structure of anti-CRISPRs, AcrF1 and AcrF2, bound to type I-F crRNA-guided CRISPR surveillance complex | | Descriptor: | Anti-CRISPR protein 30, Anti-CRISPR protein Acr30-35, CRISPR RNA (60-MER), ... | | Authors: | Chowdhury, S, Carter, J, Rollins, M.F, Jackson, R.N, Hoffmann, C, Nosaka, L, Bondy-Denomy, J, Maxwell, K.L, Davidson, A.R, Fischer, E.R, Lander, G.C, Wiedenheft, B. | | Deposit date: | 2017-02-25 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex.
Cell, 169, 2017
|
|
2LSM
 
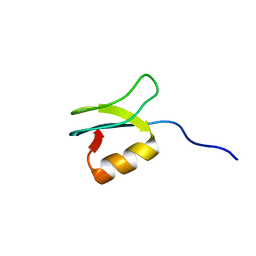 | | Solution structure of gpFI C-terminal domain | | Descriptor: | DNA-packaging protein FI | | Authors: | Popovic, A, Wu, B, Edwards, A.M, Davidson, A.R, Maxwell, K.L. | | Deposit date: | 2012-05-02 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and biochemical characterization of phage lambda FI protein (gpFI) reveals a novel mechanism of DNA packaging chaperone activity.
J.Biol.Chem., 287, 2012
|
|
2LW5
 
 | |
8F3K
 
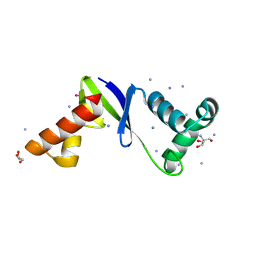 | | Anti-CRISPR protein AcrIIC5 inhibits CRISPR-Cas9 by acting as a DNA mimic | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACRIIC5Nch, AMMONIUM ION, ... | | Authors: | Shah, M, Sungwon, H, Davidson, A.R, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anti-CRISPR Protein AcrIIC5 Inhibits CRISPR-Cas9 by Occupying the Target DNA Binding Pocket.
J.Mol.Biol., 435, 2023
|
|
2K4Q
 
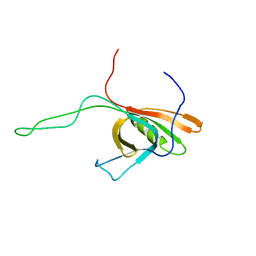 | | The Solution Structure of gpV, the Major Tail Protein from Bacteriophage Lambda | | Descriptor: | Major tail protein V | | Authors: | Pell, L.G, Kanelis, V, Howell, P, Davidson, A.R. | | Deposit date: | 2008-06-16 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2KX4
 
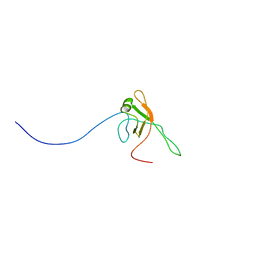 | | Solution structure of Bacteriophage Lambda gpFII | | Descriptor: | Tail attachment protein | | Authors: | Maxwell, K.L, Cardarelli, L, Neudecker, P, Davidson, A.R, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2010-04-26 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Phages have adapted the same protein fold to fulfill multiple functions in virion assembly.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2KYM
 
 | |
