2KDQ
 
 | |
2KX5
 
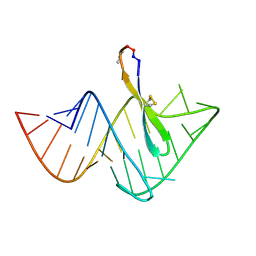 | | Recognition of HIV TAR RNA by peptide mimetic of Tat protein | | Descriptor: | Cyclic peptide mimetic of Tat protein, HIV TAR RNA | | Authors: | Davidson, A, Patora-Komisarska, K, Robinson, J, Varani, G. | | Deposit date: | 2010-04-26 | | Release date: | 2010-07-07 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Essential structural requirements for specific recognition of HIV TAR RNA by peptide mimetics of Tat protein.
Nucleic Acids Res., 39, 2011
|
|
2L8H
 
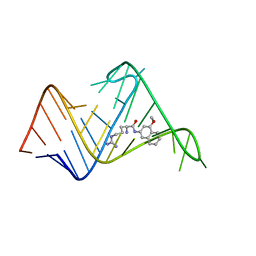 | | Chemical probe bound to HIV TAR RNA | | Descriptor: | 4-methoxynaphthalen-2-amine, ARGININE, HIV TAR RNA | | Authors: | Davidson, A, Begley, D, Lau, C, Varani, G. | | Deposit date: | 2011-01-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Small-Molecule Probe Induces a Conformation in HIV TAR RNA Capable of Binding Drug-Like Fragments.
J.Mol.Biol., 410, 2011
|
|
3CQT
 
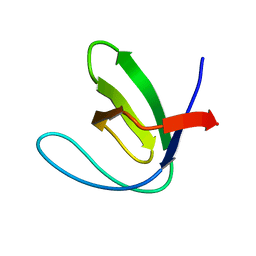 | | N53I V55L MUTANT of FYN SH3 DOMAIN | | Descriptor: | Proto-oncogene tyrosine-protein kinase Fyn | | Authors: | Neculai, A.M, Zarrine-Afsar, A, Howell, P.L, Davidson, A, Chan, H.S. | | Deposit date: | 2008-04-03 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Theoretical and experimental demonstration of the importance of specific nonnative interactions in protein folding.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1HYW
 
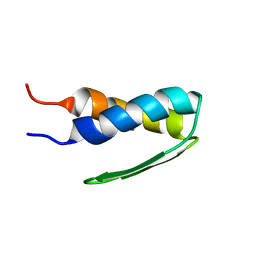 | | SOLUTION STRUCTURE OF BACTERIOPHAGE LAMBDA GPW | | Descriptor: | HEAD-TO-TAIL JOINING PROTEIN W | | Authors: | Maxwell, K.L, Yee, A.A, Booth, V, Arrowsmith, C.H, Gold, M, Davidson, A.R. | | Deposit date: | 2001-01-22 | | Release date: | 2001-04-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of bacteriophage lambda protein W, a small morphogenetic protein possessing a novel fold.
J.Mol.Biol., 308, 2001
|
|
1Z1Z
 
 | |
3FH6
 
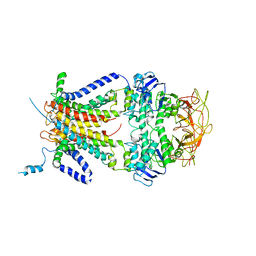 | | Crystal structure of the resting state maltose transporter from E. coli | | Descriptor: | Maltose transport system permease protein malF, Maltose transport system permease protein malG, Maltose/maltodextrin import ATP-binding protein malK | | Authors: | Khare, D, Oldham, M.L, Orelle, C, Davidson, A.L, Chen, J. | | Deposit date: | 2008-12-08 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Alternating access in maltose transporter mediated by rigid-body rotations.
Mol.Cell, 33, 2009
|
|
4JBW
 
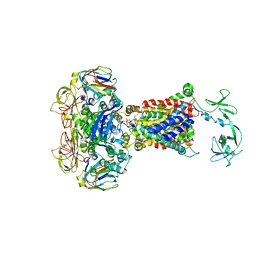 | | Crystal structure of E. coli maltose transporter MalFGK2 in complex with its regulatory protein EIIAglc | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Glucose-specific phosphotransferase enzyme IIA component, Maltose transport system permease protein MalF, ... | | Authors: | Chen, S, Oldham, M.L, Davidson, A.L, Chen, J. | | Deposit date: | 2013-02-20 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.913 Å) | | Cite: | Carbon catabolite repression of the maltose transporter revealed by X-ray crystallography.
Nature, 499, 2013
|
|
5UZ9
 
 | | Cryo EM structure of anti-CRISPRs, AcrF1 and AcrF2, bound to type I-F crRNA-guided CRISPR surveillance complex | | Descriptor: | Anti-CRISPR protein 30, Anti-CRISPR protein Acr30-35, CRISPR RNA (60-MER), ... | | Authors: | Chowdhury, S, Carter, J, Rollins, M.F, Jackson, R.N, Hoffmann, C, Nosaka, L, Bondy-Denomy, J, Maxwell, K.L, Davidson, A.R, Fischer, E.R, Lander, G.C, Wiedenheft, B. | | Deposit date: | 2017-02-25 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex.
Cell, 169, 2017
|
|
2WZQ
 
 | | Insertion Mutant E173GP174 of the NS3 protease-helicase from dengue virus | | Descriptor: | CHLORIDE ION, GLYCEROL, NS3 PROTEASE-HELICASE | | Authors: | Luo, D, Wei, N, Doan, D, Paradkar, P, Chong, Y, Davidson, A, Kotaka, M, Lescar, J, Vasudevan, S. | | Deposit date: | 2009-12-02 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Flexibility between the Protease and Helicase Domains of the Dengue Virus Ns3 Protein Conferred by the Linker Region and its Functional Implications.
J.Biol.Chem., 285, 2010
|
|
6ARZ
 
 | | Structure of a phage anti-CRISPR protein | | Descriptor: | BROMIDE ION, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Calmettes, C, Shah, M, Pawluk, A, Davidson, A.R, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Disabling a Type I-E CRISPR-Cas Nuclease with a Bacteriophage-Encoded Anti-CRISPR Protein.
MBio, 8, 2017
|
|
2RPN
 
 | | A crucial role for high intrinsic specificity in the function of yeast SH3 domains | | Descriptor: | Actin-binding protein, Actin-regulating kinase 1 | | Authors: | Stollar, E.J, Garcia, B, Chong, A, Forman-Kay, J, Davidson, A. | | Deposit date: | 2008-06-12 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural, functional, and bioinformatic studies demonstrate the crucial role of an extended peptide binding site for the SH3 domain of yeast Abp1p
J.Biol.Chem., 284, 2009
|
|
6AS4
 
 | | Structure of a phage anti-CRISPR protein | | Descriptor: | NHis AcrE1 anti-crispr protein | | Authors: | Shah, M, Calmettes, C, Pawluk, A, Mejdani, M, Davidson, A.R, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2017-08-23 | | Release date: | 2017-12-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disabling a Type I-E CRISPR-Cas Nuclease with a Bacteriophage-Encoded Anti-CRISPR Protein.
MBio, 8, 2017
|
|
6AS3
 
 | | Structure of a phage anti-CRISPR protein | | Descriptor: | NHis AcrE1 protein | | Authors: | Shah, M, Calmettes, C, Pawluk, A, Mejdani, M, Davidson, A.R, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disabling a Type I-E CRISPR-Cas Nuclease with a Bacteriophage-Encoded Anti-CRISPR Protein.
MBio, 8, 2017
|
|
2R6G
 
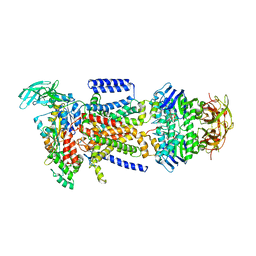 | | The Crystal Structure of the E. coli Maltose Transporter | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Maltose transport system permease protein malF, Maltose transport system permease protein malG, ... | | Authors: | Oldham, M.L, Khare, D, Quiocho, F.A, Davidson, A.L, Chen, J. | | Deposit date: | 2007-09-05 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a catalytic intermediate of the maltose transporter.
Nature, 450, 2007
|
|
7KIX
 
 | |
8F3K
 
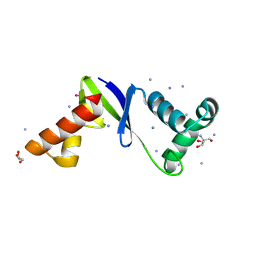 | | Anti-CRISPR protein AcrIIC5 inhibits CRISPR-Cas9 by acting as a DNA mimic | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACRIIC5Nch, AMMONIUM ION, ... | | Authors: | Shah, M, Sungwon, H, Davidson, A.R, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anti-CRISPR Protein AcrIIC5 Inhibits CRISPR-Cas9 by Occupying the Target DNA Binding Pocket.
J.Mol.Biol., 435, 2023
|
|
3FZB
 
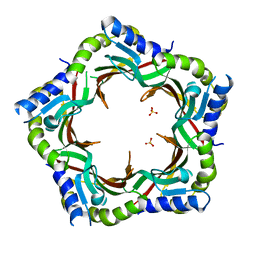 | | Crystal structure of the tail terminator protein from phage lambda (gpU-WT) | | Descriptor: | Minor tail protein U, SULFATE ION | | Authors: | Pell, L.G, Liu, A, Edmonds, E, Donaldson, L.W, Howell, P.L, Davidson, A.R. | | Deposit date: | 2009-01-24 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages.
J.Mol.Biol., 389, 2009
|
|
2AWN
 
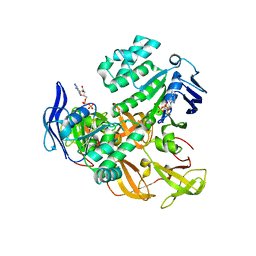 | | Crystal structure of the ADP-Mg-bound E. Coli MALK (Crystallized with ATP-Mg) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Maltose/maltodextrin import ATP-binding protein malK | | Authors: | Lu, G, Westbrooks, J.M, Davidson, A.L, Chen, J. | | Deposit date: | 2005-09-01 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ATP hydrolysis is required to reset the ATP-binding cassette dimer into the resting-state conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AWO
 
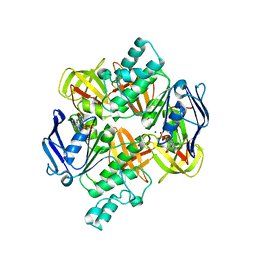 | | Crystal structure of the ADP-Mg-bound E. Coli MALK (Crystallized with ADP-Mg) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Maltose/maltodextrin import ATP-binding protein malK | | Authors: | Lu, G, Westbrooks, J.M, Davidson, A.L, Chen, J. | | Deposit date: | 2005-09-01 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | ATP hydrolysis is required to reset the ATP-binding cassette dimer into the resting-state conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1UW7
 
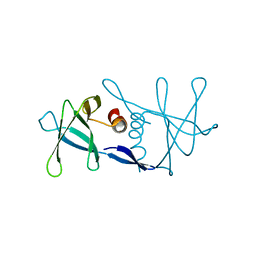 | | Nsp9 protein from SARS-coronavirus. | | Descriptor: | NSP9 | | Authors: | Sutton, G, Fry, E, Carter, L, Sainsbury, S, Walter, T, Nettleship, J, Berrow, N, Owens, R, Gilbert, R, Davidson, A, Siddell, S, Poon, L.L.M, Diprose, J, Alderton, D, Walsh, M, Grimes, J.M, Stuart, D.I. | | Deposit date: | 2004-01-30 | | Release date: | 2004-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Nsp9 Replicase Protein of Sars-Coronavirus, Structure and Functional Insights
Structure, 12, 2004
|
|
3FZ2
 
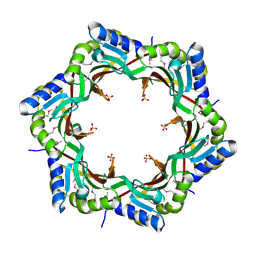 | | Crystal structure of the tail terminator protein from phage lambda (gpU-D74A) | | Descriptor: | Minor tail protein U, SULFATE ION | | Authors: | Pell, L.G, Liu, A, Edmonds, E, Donaldson, L.W, Howell, P.L, Davidson, A.R. | | Deposit date: | 2009-01-23 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages.
J.Mol.Biol., 389, 2009
|
|
1Q1E
 
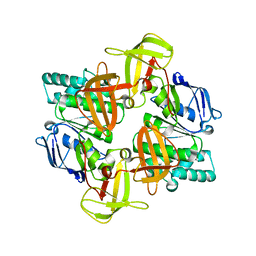 | | The ATPase component of E. coli maltose transporter (MalK) in the nucleotide-free form | | Descriptor: | Maltose/maltodextrin transport ATP-binding protein malK | | Authors: | Chen, J, Lu, G, Lin, J, Davidson, A.L, Quiocho, F.A. | | Deposit date: | 2003-07-19 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle
Mol.Cell, 12, 2003
|
|
2LCS
 
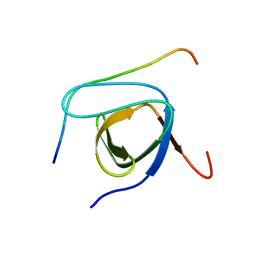 | | Yeast Nbp2p SH3 domain in complex with a peptide from Ste20p | | Descriptor: | NAP1-binding protein 2, Serine/threonine-protein kinase STE20 | | Authors: | Gorelik, M, Davidson, A.R. | | Deposit date: | 2011-05-07 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Distinct Peptide Binding Specificities of Src Homology 3 (SH3) Protein Domains Can Be Determined by Modulation of Local Energetics across the Binding Interface.
J.Biol.Chem., 287, 2012
|
|
2LTF
 
 | |
