8D2K
 
 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Cleavage Intermediate 2) | | 分子名称: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*AP*GP*A)-3'), ... | | 著者 | Rai, J, Das, A, Li, H. | | 登録日 | 2022-05-30 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.43 Å) | | 主引用文献 | Coupled catalytic states and the role of metal coordination in Cas9.
Nat Catal, 6, 2023
|
|
8D2O
 
 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Post-cleavage 2) | | 分子名称: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*AP*TP*AP*CP*AP*CP*CP*AP*AP*GP*CP*T)-3'), ... | | 著者 | Rai, J, Das, A, Li, H. | | 登録日 | 2022-05-30 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Coupled catalytic states and the role of metal coordination in Cas9.
Nat Catal, 6, 2023
|
|
8D2Q
 
 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Post-cleavage 1) | | 分子名称: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*AP*TP*AP*CP*AP*CP*CP*AP*AP*GP*CP*T)-3'), ... | | 著者 | Rai, J, Das, A, Li, H. | | 登録日 | 2022-05-30 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.58 Å) | | 主引用文献 | Coupled catalytic states and the role of metal coordination in Cas9.
Nat Catal, 6, 2023
|
|
4XW6
 
 | | X-ray structure of PKAc with ADP, free phosphate ion, CP20, magnesium ions | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Gerlits, O, Tian, J, Das, A, Taylor, S, Langan, P, Heller, T.W, Kovalevsky, A. | | 登録日 | 2015-01-28 | | 公開日 | 2015-05-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Phosphoryl Transfer Reaction Snapshots in Crystals: INSIGHTS INTO THE MECHANISM OF PROTEIN KINASE A CATALYTIC SUBUNIT.
J.Biol.Chem., 290, 2015
|
|
4XW5
 
 | | X-ray structure of PKAc with ATP, CP20, calcium ions | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | 著者 | Gerlits, O, Tian, J, Das, A, Taylor, S, Langan, P, Heller, T.W, Kovalevsky, A. | | 登録日 | 2015-01-28 | | 公開日 | 2015-05-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Phosphoryl Transfer Reaction Snapshots in Crystals: INSIGHTS INTO THE MECHANISM OF PROTEIN KINASE A CATALYTIC SUBUNIT.
J.Biol.Chem., 290, 2015
|
|
4XW4
 
 | | X-ray structure of PKAc with AMPPNP, SP20, calcium ions | | 分子名称: | CALCIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | 著者 | Gerlits, O, Tian, J, Das, A, Taylor, S, Langan, P, Heller, T.W, Kovalevsky, A. | | 登録日 | 2015-01-28 | | 公開日 | 2015-05-06 | | 最終更新日 | 2015-07-01 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Phosphoryl Transfer Reaction Snapshots in Crystals: INSIGHTS INTO THE MECHANISM OF PROTEIN KINASE A CATALYTIC SUBUNIT.
J.Biol.Chem., 290, 2015
|
|
2WHH
 
 | | HIV-1 protease tethered dimer Q-product complex along with nucleophilic water molecule | | 分子名称: | GLUTAMIC ACID, PARA-NITROPHENYLALANINE, POL PROTEIN | | 著者 | Prashar, V, Bihani, S, Das, A, Ferrer, J.L, Hosur, M.V. | | 登録日 | 2009-05-05 | | 公開日 | 2009-12-01 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Catalytic Water Co-Existing with a Product Peptide in the Active Site of HIV-1 Protease Revealed by X- Ray Structure Analysis.
Plos One, 4, 2009
|
|
6LFH
 
 | |
3KT2
 
 | | Crystal Structure of N88D mutant HIV-1 Protease | | 分子名称: | Protease | | 著者 | Bihani, S.C, Das, A, Prashar, V, Ferrer, J.L, Hosur, M.V. | | 登録日 | 2009-11-24 | | 公開日 | 2010-02-16 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.651 Å) | | 主引用文献 | Resistance mechanism revealed by crystal structures of unliganded nelfinavir-resistant HIV-1 protease non-active site mutants N88D and N88S.
Biochem.Biophys.Res.Commun., 389, 2009
|
|
3KT5
 
 | | Crystal Structure of N88S mutant HIV-1 Protease | | 分子名称: | Protease | | 著者 | Bihani, S.C, Das, A, Prashar, V, Ferrer, J.L, Hosur, M.V. | | 登録日 | 2009-11-24 | | 公開日 | 2010-02-16 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Resistance mechanism revealed by crystal structures of unliganded nelfinavir-resistant HIV-1 protease non-active site mutants N88D and N88S.
Biochem.Biophys.Res.Commun., 389, 2009
|
|
1JO1
 
 | | N7-Guanine Adduct of 2,7-diaminomitosene with DNA | | 分子名称: | 5'-D(*GP*TP*GP*(DAJ)GP*TP*AP*TP*AP*CP*CP*AP*C)-3', DECARBAMOYL-2,7-DIAMINOMITOSENE | | 著者 | Subramaniam, G, Paz, M.M, Kumar, G.S, Das, A, Palom, Y, Clement, C.C, Patel, D.J, Tomasz, M. | | 登録日 | 2001-07-26 | | 公開日 | 2001-09-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a guanine-N7-linked complex of the mitomycin C metabolite 2,7-diaminomitosene and DNA. Basis of sequence selectivity.
Biochemistry, 40, 2001
|
|
3DOX
 
 | | X-ray structure of HIV-1 protease in situ product complex | | 分子名称: | A PEPTIDE SUBSTRATE-PIV, A PEPTIDE SUBSTRATE-SQNY, HIV-1 PROTEASE | | 著者 | Hosur, M.V, Ferrer, J.-L, Das, A, Prashar, V, Bihani, S. | | 登録日 | 2008-07-07 | | 公開日 | 2008-09-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray structure of HIV-1 protease in situ product complex
Proteins, 74, 2009
|
|
3N3I
 
 | | Crystal Structure of G48V/C95F tethered HIV-1 Protease/Saquinavir complex | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Protease | | 著者 | Prashar, V, Bihani, S.C, Das, A, Rao, D.R, Hosur, M.V. | | 登録日 | 2010-05-20 | | 公開日 | 2010-06-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Insights into the mechanism of drug resistance: X-ray structure analysis of G48V/C95F tethered HIV-1 protease dimer/saquinavir complex
Biochem.Biophys.Res.Commun., 396, 2010
|
|
2NPH
 
 | |
3H4C
 
 | | Structure of the C-terminal Domain of Transcription Factor IIB from Trypanosoma brucei | | 分子名称: | 1,2-ETHANEDIOL, Transcription factor TFIIB-like | | 著者 | Syed Ibrahim, B, Kanneganti, N, Rieckhof, G.E, Das, A, Laurents, D.V, Palenchar, J.B, Bellofatto, V, Wah, D.A. | | 登録日 | 2009-04-18 | | 公開日 | 2009-08-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the C-terminal domain of transcription factor IIB from Trypanosoma brucei.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1Y80
 
 | | Structure of a corrinoid (factor IIIm)-binding protein from Moorella thermoacetica | | 分子名称: | CO-5-METHOXYBENZIMIDAZOLYLCOBAMIDE, Predicted cobalamin binding protein, UNKNOWN ATOM OR ION | | 著者 | Liu, Z.-J, Fu, Z.-Q, Tempel, W, Das, A, Habel, J, Zhou, W, Chang, J, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Ljungdahl, L, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2004-12-10 | | 公開日 | 2005-01-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of a corrinoid (factor IIIm)-binding protein from Moorella thermoacetica
To be published
|
|
8OR1
 
 | | Co-crystal strucutre of PD-L1 with low molecular weight inhibitor | | 分子名称: | 5-[[5-[[2-chloranyl-3-(2-fluorophenyl)phenyl]methoxy]-2-[(~{E})-2-hydroxyethyliminomethyl]phenoxy]methyl]pyridine-3-carbonitrile, Programmed cell death 1 ligand 1 | | 著者 | Zhang, H, Zhou, S, Wu, C, Zhu, M, Yu, Q, Wang, X, Awadasseid, A, Plewka, J, Magiera-Mularz, K, Wu, Y, Zhang, W. | | 登録日 | 2023-04-12 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Design, Synthesis, and Antitumor Activity Evaluation of 2-Arylmethoxy-4-(2,2'-dihalogen-substituted biphenyl-3-ylmethoxy) Benzylamine Derivatives as Potent PD-1/PD-L1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
6EKA
 
 | | Solid-state MAS NMR structure of the HELLF prion amyloid fibrils | | 分子名称: | Podospora anserina S mat+ genomic DNA chromosome 3, supercontig 2 | | 著者 | Martinez, D, Daskalov, A, Andreas, L, Bardiaux, B, Coustou, V, Stanek, J, Berbon, M, Noubhani, M, Kauffmann, B, Wall, J.S, Pintacuda, G, Saupe, S.J, Habenstein, B, Loquet, A. | | 登録日 | 2017-09-25 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Structural and molecular basis of cross-seeding barriers in amyloids
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7L7H
 
 | | Alpha-synuclein fibrils | | 分子名称: | Alpha-synuclein | | 著者 | Hojjatian, A, Dasari, A. | | 登録日 | 2020-12-28 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Distinct cryo-EM Structure of Alpha-synuclein Filaments derived by Tau
To Be Published
|
|
7A8P
 
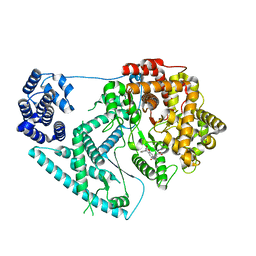 | | Structure of human mitochondrial RNA polymerase in complex with IMT inhibitor. | | 分子名称: | (3~{R})-1-[(2~{R})-2-[4-(2-chloranyl-4-fluoranyl-phenyl)-2-oxidanylidene-chromen-7-yl]oxypropanoyl]piperidine-3-carboxylic acid, DNA-directed RNA polymerase, mitochondrial | | 著者 | Hillen, H.S, Bonekamp, N, Peter, B, Felser, A, Bergbrede, T, Choidas, A, Horn, M, Unger, A, di Lucrezia, R, Atanassov, I, Li, X, Koch, U, Menninger, S, Boros, J, Habenberger, P, Giavalisco, P, Cramer, P, Denzel, M, Nussbaumer, P, Klebl, B, Falkenberg, M, Gustafsson, C.M, Larsson, N.G. | | 登録日 | 2020-08-30 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Small-molecule inhibitors of human mitochondrial DNA transcription.
Nature, 588, 2020
|
|
5SVW
 
 | |
5SVU
 
 | |
5SVG
 
 | |
5SVV
 
 | |
4XWH
 
 | | Crystal structure of the human N-acetyl-alpha-glucosaminidase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-N-acetylglucosaminidase, ... | | 著者 | Birrane, G, Meiyappan, M, Dassier, A. | | 登録日 | 2015-01-28 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structural characterization of the alpha-N-acetylglucosaminidase, a key enzyme in the pathogenesis of Sanfilippo syndrome B.
J.Struct.Biol., 205, 2019
|
|
