7XL4
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (closed lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7XL3
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (open lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7VF9
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7X89
 
 | | Tid1 | | Descriptor: | DnaJ homolog subfamily A member 3, mitochondrial | | Authors: | Jang, J, Lee, S.H, Kang, D.H, Sim, D.W, Jo, K.S, Ryu, H, Kim, E.H, Ryu, K.S, Lee, J.H, Kim, J.H, Won, H.S. | | Deposit date: | 2022-03-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies on the J-domain and GF-motif of the mitochondrial Hsp40, Tid1
To Be Published
|
|
1RQ2
 
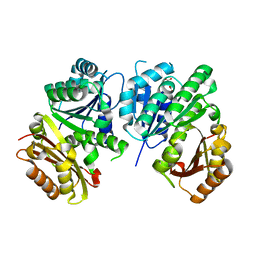 | | MYCOBACTERIUM TUBERCULOSIS FTSZ IN COMPLEX WITH CITRATE | | Descriptor: | CITRIC ACID, Cell division protein ftsZ | | Authors: | Leung, A.K.W, White, E.L, Ross, L.J, Reynolds, R.C, DeVito, J.A, Borhani, D.W. | | Deposit date: | 2003-12-04 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches.
J.Mol.Biol., 342, 2004
|
|
1RLU
 
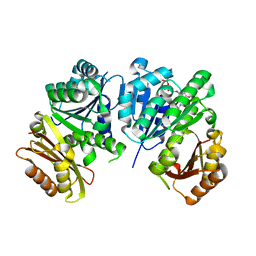 | | Mycobacterium tuberculosis FtsZ in complex with GTP-gamma-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell division protein ftsZ, GLYCEROL | | Authors: | Leung, A.K.W, White, E.L, Ross, L.J, Reynolds, R.C, DeVito, J.A, Borhani, D.W. | | Deposit date: | 2003-11-26 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches.
J.Mol.Biol., 342, 2004
|
|
1SJJ
 
 | |
1RQ7
 
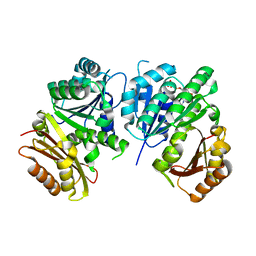 | | MYCOBACTERIUM TUBERCULOSIS FTSZ IN COMPLEX WITH GDP | | Descriptor: | Cell division protein ftsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Leung, A.K.W, White, E.L, Ross, L.J, Reynolds, R.C, DeVito, J.A, Borhani, D.W. | | Deposit date: | 2003-12-04 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches.
J.Mol.Biol., 342, 2004
|
|
4CAA
 
 | | CLEAVED ANTICHYMOTRYPSIN T345R | | Descriptor: | ANTICHYMOTRYPSIN | | Authors: | Lukacs, C.M, Christianson, D.W. | | Deposit date: | 1997-08-14 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering an anion-binding cavity in antichymotrypsin modulates the "spring-loaded" serpin-protease interaction.
Biochemistry, 37, 1998
|
|
1HRD
 
 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Britton, K.L, Baker, P.J, Stillman, T.J, Rice, D.W. | | Deposit date: | 1996-04-03 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The structure of Pyrococcus furiosus glutamate dehydrogenase reveals a key role for ion-pair networks in maintaining enzyme stability at extreme temperatures.
Structure, 3, 1995
|
|
1HQ5
 
 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH S-(2-BORONOETHYL)-L-CYSTEINE, AN L-ARGININE ANALOGUE | | Descriptor: | ARGINASE 1, MANGANESE (II) ION, S-2-(BORONOETHYL)-L-CYSTEINE | | Authors: | Kim, N.N, Cox, J.D, Baggio, R.F, Emig, F.A, Mistry, S.K, Harper, S.L, Speicher, D.W, Morris Jr, S.M, Ash, D.E, Traish, A, Christianson, D.W. | | Deposit date: | 2000-12-14 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing erectile function: S-(2-boronoethyl)-L-cysteine binds to arginase as a transition state analogue and enhances smooth muscle relaxation in human penile corpus cavernosum.
Biochemistry, 40, 2001
|
|
1GLF
 
 | | CRYSTAL STRUCTURES OF ESCHERICHIA COLI GLYCEROL KINASE AND THE MUTANT A65T IN AN INACTIVE TETRAMER: CONFORMATIONAL CHANGES AND IMPLICATIONS FOR ALLOSTERIC REGULATION | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Feese, M.D, Faber, H.R, Bystrom, C.E, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1998-08-30 | | Release date: | 1998-10-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Glycerol kinase from Escherichia coli and an Ala65-->Thr mutant: the crystal structures reveal conformational changes with implications for allosteric regulation.
Structure, 6, 1998
|
|
1BU6
 
 | | CRYSTAL STRUCTURES OF ESCHERICHIA COLI GLYCEROL KINASE AND THE MUTANT A65T IN AN INACTIVE TETRAMER: CONFORMATIONAL CHANGES AND IMPLICATIONS FOR ALLOSTERIC REGULATION | | Descriptor: | GLYCEROL, PROTEIN (GLYCEROL KINASE), SULFATE ION | | Authors: | Feese, M.D, Faber, H.R, Bystrom, C.E, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1998-08-30 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Glycerol kinase from Escherichia coli and an Ala65-->Thr mutant: the crystal structures reveal conformational changes with implications for allosteric regulation.
Structure, 6, 1998
|
|
3ZNC
 
 | | MURINE CARBONIC ANHYDRASE IV COMPLEXED WITH BRINZOLAMIDE | | Descriptor: | (+)-4-ETHYLAMINO-3,4-DIHYDRO-2-(METHOXY)PROPYL-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE IV, ZINC ION | | Authors: | Stams, T, Chen, Y, Christianson, D.W. | | Deposit date: | 1998-02-10 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of murine carbonic anhydrase IV and human carbonic anhydrase II complexed with brinzolamide: molecular basis of isozyme-drug discrimination.
Protein Sci., 7, 1998
|
|
1TIM
 
 | | STRUCTURE OF TRIOSE PHOSPHATE ISOMERASE FROM CHICKEN MUSCLE | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Banner, D.W, Bloomer, A.C, Petsko, G.A, Phillips, D.C, Wilson, I.A. | | Deposit date: | 1976-09-01 | | Release date: | 1976-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic coordinates for triose phosphate isomerase from chicken muscle.
Biochem.Biophys.Res.Commun., 72, 1976
|
|
7NPP
 
 | |
7NY3
 
 | |
7PTD
 
 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C MUTANT R163K | | Descriptor: | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Probing the roles of active site residues in phosphatidylinositol-specific phospholipase C from Bacillus cereus by site-directed mutagenesis.
Biochemistry, 36, 1997
|
|
7PQ0
 
 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) C94S inactive mutant in complex with human eIF4A - Crystal form B | | Descriptor: | Burkholderia Lethal Factor 1 (BLF1), Eukaryotic initiation factor 4A-I | | Authors: | Mobbs, G.W, Aziz, A.A, Dix, S.R, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2021-09-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
7PPZ
 
 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) C94S inactive mutant in complex with human eIF4A - Crystal form A | | Descriptor: | Burkholderia Lethal Factor 1 (BLF1), Eukaryotic initiation factor 4A-I | | Authors: | Mobbs, G.W, Aziz, A.A, Dix, S.R, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2021-09-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
3TU8
 
 | | Crystal Structure of the Burkholderia Lethal Factor 1 (BLF1) | | Descriptor: | BROMIDE ION, Burkholderia Lethal Factor 1 (BLF1) | | Authors: | Cruz, A, Hautbergue, G.M, Artymiuk, P.J, Baker, P.J, Chang, C.T, Mahadi, N.M, Mobbs, G.W, Mohamed, R, Nathan, S, Partridge, L.J, Raih, M.F, Ruzheinikov, S.N, Sedelnikova, S.E, Wilson, S.A, Rice, D.W. | | Deposit date: | 2011-09-16 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A Burkholderia pseudomallei toxin inhibits helicase activity of translation factor eIF4A.
Science, 334, 2011
|
|
3NH9
 
 | | Nucleotide Binding Domain of Human ABCB6 (ATP bound structure) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family B member 6, mitochondrial, ... | | Authors: | Haffke, M, Menzel, A, Carius, Y, Jahn, D, Heinz, D.W. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the nucleotide-binding domain of the human ABCB6 transporter and its complexes with nucleotides.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3TUA
 
 | | Crystal Structure of the Burkholderia Lethal Factor 1 (BLF1) C94S mutant | | Descriptor: | Burkholderia Lethal Factor 1 (BLF1) | | Authors: | Cruz, A, Hautbergue, G.M, Artymiuk, P.J, Baker, P.J, Chang, C.T, Mahadi, N.M, Mobbs, G.W, Mohamed, R, Nathan, S, Partridge, L.J, Raih, M.F, Ruzheinikov, S.N, Sedelnikova, S.E, Wilson, S.A, Rice, D.W. | | Deposit date: | 2011-09-16 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | A Burkholderia pseudomallei toxin inhibits helicase activity of translation factor eIF4A.
Science, 334, 2011
|
|
3NJ8
 
 | |
5PTD
 
 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C MUTANT H32A | | Descriptor: | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W. | | Deposit date: | 1997-07-18 | | Release date: | 1998-01-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Probing the roles of active site residues in phosphatidylinositol-specific phospholipase C from Bacillus cereus by site-directed mutagenesis.
Biochemistry, 36, 1997
|
|
