7T5G
 
 | | Structure of rabies virus phosphoprotein C-terminal domain, S210E mutant | | Descriptor: | Phosphoprotein, SULFATE ION | | Authors: | Zhan, J, Metcalfe, R.D, Gooley, P.R, Griffin, M.D.W. | | Deposit date: | 2021-12-12 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis of Functional Effects of Phosphorylation of the C-Terminal Domain of the Rabies Virus P Protein.
J.Virol., 96, 2022
|
|
9B3T
 
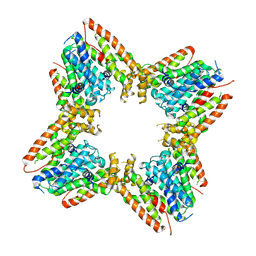 | |
9CBK
 
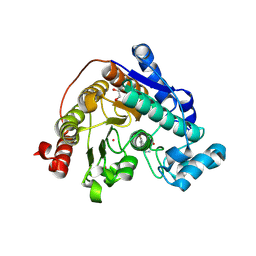 | | Crystal Structure of Danio rerio Histone Deacetylase 6 in Complex with p-Aminomethyl Phenylthioketone | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(aminomethyl)phenyl]-2-sulfanylethan-1-one, Hdac6 protein, ... | | Authors: | Goulart Stollmaier, J, Watson, P.R, Christianson, D.W. | | Deposit date: | 2024-06-19 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Design, Synthesis, and Structural Evaluation of Acetylated Phenylthioketone Inhibitors of HDAC10.
Acs Med.Chem.Lett., 15, 2024
|
|
9CBH
 
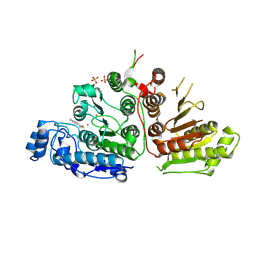 | | Crystal Structure of Danio rerio Histone Deacetylase 10 in Complex with p-Aminomethyl Phenylthioketone | | Descriptor: | 1-[4-(aminomethyl)phenyl]-2-sulfanylethan-1-one, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Goulart Stollmaier, J, Christianson, D.W. | | Deposit date: | 2024-06-19 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design, Synthesis, and Structural Evaluation of Acetylated Phenylthioketone Inhibitors of HDAC10.
Acs Med.Chem.Lett., 15, 2024
|
|
7T5H
 
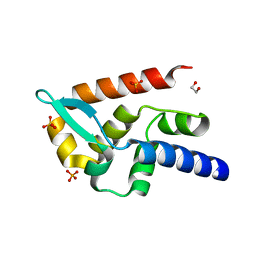 | | Structure of rabies virus phosphoprotein C-terminal domain, wild type | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Phosphoprotein, ... | | Authors: | Zhan, J, Metcalfe, R.D, Gooley, P.R, Griffin, M.D.W. | | Deposit date: | 2021-12-12 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis of Functional Effects of Phosphorylation of the C-Terminal Domain of the Rabies Virus P Protein.
J.Virol., 96, 2022
|
|
9CBJ
 
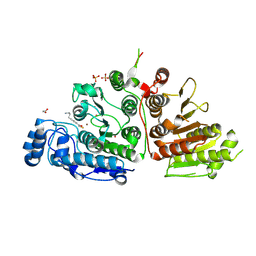 | | Crystal Structure of Danio rerio Histone Deacetylase 10 in Complex with p-Aminopropyl Phenylthioketone | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(3-aminopropyl)phenyl]-2-sulfanylethan-1-one, PHOSPHATE ION, ... | | Authors: | Goulart Stollmaier, J, Christianson, D.W. | | Deposit date: | 2024-06-19 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Design, Synthesis, and Structural Evaluation of Acetylated Phenylthioketone Inhibitors of HDAC10.
Acs Med.Chem.Lett., 15, 2024
|
|
9CBG
 
 | | Crystal Structure of Danio rerio Histone Deacetylase 10 in Complex with m-Aminoethyl Phenylthioketone | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-(2-aminoethyl)phenyl]-2-sulfanylethan-1-one, PHOSPHATE ION, ... | | Authors: | Goulart Stollmaier, J, Christianson, D.W. | | Deposit date: | 2024-06-19 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, Synthesis, and Structural Evaluation of Acetylated Phenylthioketone Inhibitors of HDAC10.
Acs Med.Chem.Lett., 15, 2024
|
|
9BUL
 
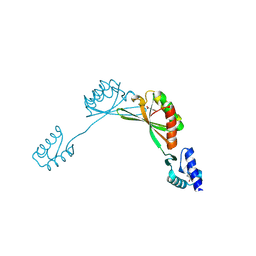 | |
9BQV
 
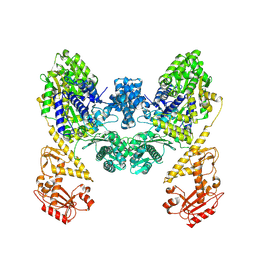 | | DdmD dimer apoprotein | | Descriptor: | Helicase/UvrB N-terminal domain-containing protein | | Authors: | Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Plasmid targeting and destruction by the DdmDE bacterial defence system.
Nature, 630, 2024
|
|
9CA2
 
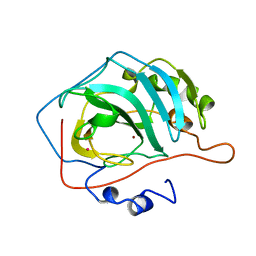 | |
6WYO
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 1 (CD1) H82F F202Y double mutant complexed with Trichostatin A | | Descriptor: | Histone deacetylase 6, POTASSIUM ION, TRICHOSTATIN A, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.30000281 Å) | | Cite: | Binding of inhibitors to active-site mutants of CD1, the enigmatic catalytic domain of histone deacetylase 6.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7SQ1
 
 | | BG505.MD39TS Env trimer in complex with Fab from antibody C05 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C05 Fab Light chain, ... | | Authors: | Moore, A, Du, J, Xu, Z, Walker, S, Kulp, D.W, Pallesen, J. | | Deposit date: | 2021-11-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Induction of tier-2 neutralizing antibodies in mice with a DNA-encoded HIV envelope native like trimer.
Nat Commun, 13, 2022
|
|
3OOC
 
 | | Crystal structure of the membrane fusion protein CusB from Escherichia coli | | Descriptor: | Cation efflux system protein cusB | | Authors: | Su, C.-C, Yang, F, Long, F, Reyon, D, Routh, M.D, Kuo, D.W, Mokhtari, A.K, Van Ornam, J.D, Rabe, K.L, Hoy, J.A, Lee, Y.J, Rajashankar, K.R, Yu, E.W. | | Deposit date: | 2010-08-30 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Crystal structure of the membrane fusion protein CusB from Escherichia coli.
J.Mol.Biol., 393, 2009
|
|
6WSJ
 
 | |
5GZ0
 
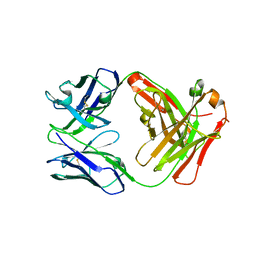 | | Crystal structure of FM329, a recombinant Fab adopted from cetuximab | | Descriptor: | FM329 heavy chain, FM329 light chain | | Authors: | Sim, D.W, Kim, J.H, Kim, Y.P, Won, H.S. | | Deposit date: | 2016-09-26 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of FM329, a recombinant Fab adopted from cetuximab
To Be Published
|
|
3P7O
 
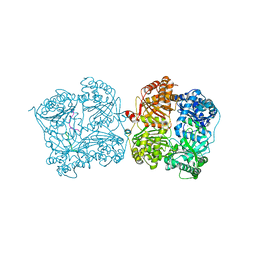 | |
3ONM
 
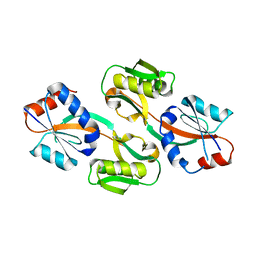 | | Effector binding Domain of LysR-Type transcription factor RovM from Y. pseudotuberculosis | | Descriptor: | Transcriptional regulator LrhA | | Authors: | Quade, N, Diekmann, M, Haffke, M, Heroven, A.K, Dersch, P, Heinz, D.W. | | Deposit date: | 2010-08-30 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the effector-binding domain of the LysR-type transcription factor RovM from Yersinia pseudotuberculosis.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5H1W
 
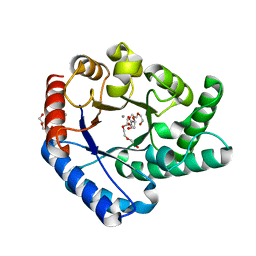 | | Crystal Structure of Hyperthermophilic Thermotoga maritima L-Ketose-3-Epimerase with Mn2+ and L(+)-Erythrulose | | Descriptor: | L-Erythrulose, MANGANESE (II) ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Cao, T.P, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2016-10-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | TM0416, a Hyperthermophilic Promiscuous Nonphosphorylated Sugar Isomerase, Catalyzes Various C5and C6Epimerization Reactions
Appl. Environ. Microbiol., 83, 2017
|
|
6XB0
 
 | |
6XQT
 
 | | Room-temperature X-ray Crystal structure of SARS-CoV-2 main protease in complex with Narlaprevir | | Descriptor: | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropylamino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kneller, D.W, Kovalevsky, A, Coates, L. | | Deposit date: | 2020-07-10 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Malleability of the SARS-CoV-2 3CL M pro Active-Site Cavity Facilitates Binding of Clinical Antivirals.
Structure, 28, 2020
|
|
3OM6
 
 | | Crystal structure of B. megaterium levansucrase mutant Y247A | | Descriptor: | CALCIUM ION, Levansucrase, SULFATE ION, ... | | Authors: | Strube, C.P, Homann, A, Gamer, M, Jahn, D, Seibel, J, Heinz, D.W. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Polysaccharide Synthesis of the Levansucrase SacB from Bacillus megaterium Is Controlled by Distinct Surface Motifs.
J.Biol.Chem., 286, 2011
|
|
3OM7
 
 | | Crystal structure of B. megaterium levansucrase mutant Y247W | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Levansucrase, ... | | Authors: | Strube, C.P, Homann, A, Gamer, M, Jahn, D, Seibel, J, Heinz, D.W. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Polysaccharide Synthesis of the Levansucrase SacB from Bacillus megaterium Is Controlled by Distinct Surface Motifs.
J.Biol.Chem., 286, 2011
|
|
7TEH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TFR
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with NBH-2 | | Descriptor: | (1R,2S,5S)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TDU
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-1 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxo(1-~2~H)pyrrolidin-3-yl]propan-2-yl}-3-{N-[tert-butyl(~2~H)carbamoyl]-3-methyl-L-(N-~2~H)valyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-(~2~H)carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
