2D2H
 
 | | OpdA from Agrobacterium radiobacter with bound inhibitor trimethyl phosphate at 1.8 A resolution | | Descriptor: | COBALT (II) ION, TRIMETHYL PHOSPHATE, phosphotriesterase | | Authors: | Jackson, C, Kim, H.K, Carr, P.D, Liu, J.W, Ollis, D.L. | | Deposit date: | 2005-09-09 | | Release date: | 2005-09-20 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of an enzyme-product complex reveals the critical role of a terminal hydroxide nucleophile in the bacterial phosphotriesterase mechanism
Biochim.Biophys.Acta, 1752, 2005
|
|
2EF9
 
 | | Structural and mechanistic changes along an engineered path from metallo to non-metallo KDO8P synthase | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, ARABINOSE-5-PHOSPHATE, PHOSPHOENOLPYRUVATE | | Authors: | Kona, F, Xu, X, Martin, P, Kuzmic, P, Gatti, D.L. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Changes along an Engineered Path from Metallo to Nonmetallo 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthases
Biochemistry, 46, 2007
|
|
2D2J
 
 | | OpdA from Agrobacterium radiobacter without inhibitor/product present at 1.75 A resolution | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, phosphotriesterase | | Authors: | Jackson, C, Kim, H.K, Carr, P.D, Liu, J.W, Ollis, D.L. | | Deposit date: | 2005-09-09 | | Release date: | 2005-09-20 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of an enzyme-product complex reveals the critical role of a terminal hydroxide nucleophile in the bacterial phosphotriesterase mechanism
Biochim.Biophys.Acta, 1752, 2005
|
|
2D2G
 
 | | OpdA from Agrobacterium radiobacter with bound product dimethylthiophosphate | | Descriptor: | COBALT (II) ION, O,O-DIMETHYL HYDROGEN THIOPHOSPHATE, phosphotriesterase | | Authors: | Jackson, C, Kim, H.K, Carr, P.D, Liu, J.W, Ollis, D.L. | | Deposit date: | 2005-09-08 | | Release date: | 2005-09-20 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of an enzyme-product complex reveals the critical role of a terminal hydroxide nucleophile in the bacterial phosphotriesterase mechanism
Biochim.Biophys.Acta, 1752, 2005
|
|
2EST
 
 | |
2FRC
 
 | | CYTOCHROME C (REDUCED) FROM EQUUS CABALLUS, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Qi, P.X, Di Stefano, D.L, Wand, A.J. | | Deposit date: | 1996-07-02 | | Release date: | 1997-07-29 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of horse heart ferricytochrome c and detection of redox-related structural changes by high-resolution 1H NMR.
Biochemistry, 35, 1996
|
|
2FSU
 
 | | Crystal Structure of the PhnH Protein from Escherichia Coli | | Descriptor: | ACETATE ION, Protein phnH, SODIUM ION | | Authors: | Adams, M.A, Luo, Y, Zechel, D.L, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-23 | | Release date: | 2007-03-27 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of PhnH: an essential component of carbon-phosphorus lyase in Escherichia coli.
J.Bacteriol., 190, 2008
|
|
2GYS
 
 | | 2.7 A structure of the extracellular domains of the human beta common receptor involved in IL-3, IL-5, and GM-CSF signalling | | Descriptor: | Cytokine receptor common beta chain, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Carr, P.D, Conlan, F, Ford, S, Ollis, D.L, Young, I.G. | | Deposit date: | 2006-05-09 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An improved resolution structure of the human beta common receptor involved in IL-3, IL-5 and GM-CSF signalling which gives better definition of the high-affinity binding epitope.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2H7X
 
 | | Pikromycin Thioesterase adduct with reduced triketide affinity label | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Giraldes, J.W, Akey, D.L, Kittendorf, J.D, Sherman, D.H, Smith, J.S, Fecik, R.A. | | Deposit date: | 2006-06-05 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Mechanistic Insights of Polyketide Macrolactonization from Polyketide-based Affinity Labels
NAT.CHEM.BIOL., 2, 2006
|
|
2H8N
 
 | |
2HFK
 
 | | Pikromycin thioesterase in complex with product 10-deoxymethynolide | | Descriptor: | (3R,4S,5S,7R,9E,11R,12R)-12-ETHYL-4-HYDROXY-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Akey, D.L, Kittendorf, J.D, Giraldes, J.W, Fecik, R.A, Sherman, D.H, Smith, J.L. | | Deposit date: | 2006-06-24 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Macrolactonization by the Pikromycin Thioesterase
NAT.CHEM.BIOL., 2, 2006
|
|
2GNK
 
 | | GLNK, A SIGNAL PROTEIN FROM E. COLI | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PROTEIN (NITROGEN REGULATORY PROTEIN) | | Authors: | Xu, Y, Cheah, E, Carr, P.D, van Heeswijk, W.C, Westerhoff, H.V, Vasudevan, S.G, Ollis, D.L. | | Deposit date: | 1998-07-14 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GlnK, a PII-homologue: structure reveals ATP binding site and indicates how the T-loops may be involved in molecular recognition.
J.Mol.Biol., 282, 1998
|
|
2H7Y
 
 | | Pikromycin Thioesterase with covalent affinity label | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Giraldes, J.W, Akey, D.L, Kittendorf, J.D, Sherman, D.H, Smith, J.S, Fecik, R.A. | | Deposit date: | 2006-06-06 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insights of Polyketide Macrolactonization from Polyketide-based Affinity Labels
NAT.CHEM.BIOL., 2, 2006
|
|
2HFJ
 
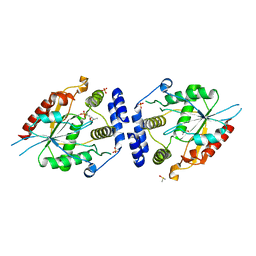 | | Pikromycin thioesterase with covalent pentaketide affinity label | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Akey, D.L, Kittendorf, J.D, Giraldes, J.W, Fecik, R.A, Sherman, D.H, Smith, J.L. | | Deposit date: | 2006-06-24 | | Release date: | 2006-09-19 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Macrolactonization by the Pikromycin Thioesterase
NAT.CHEM.BIOL., 2, 2006
|
|
2EZ6
 
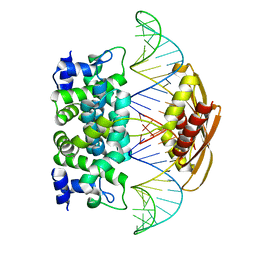 | | Crystal structure of Aquifex aeolicus RNase III (D44N) complexed with product of double-stranded RNA processing | | Descriptor: | 28-MER, MAGNESIUM ION, Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-11-10 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insight into the Mechanism of Double-Stranded RNA Processing by Ribonuclease III.
Cell(Cambridge,Mass.), 124, 2006
|
|
2I2J
 
 | | NMR structure of UA159sp in TFE | | Descriptor: | Competence stimulating peptide | | Authors: | Syvitski, R.T, Jakeman, D.L, Li, Y. | | Deposit date: | 2006-08-16 | | Release date: | 2006-10-17 | | Last modified: | 2020-03-04 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Analysis of Quorum-Sensing Signaling Peptides from Streptococcus mutans.
J.Bacteriol., 189, 2007
|
|
2I2R
 
 | | Crystal structure of the KChIP1/Kv4.3 T1 complex | | Descriptor: | CALCIUM ION, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 3, ... | | Authors: | Findeisen, F, Pioletti, M, Minor Jr, D.L. | | Deposit date: | 2006-08-16 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Three-dimensional structure of the KChIP1-Kv4.3 T1 complex reveals a cross-shaped octamer
Nat.Struct.Mol.Biol., 13, 2006
|
|
2I5N
 
 | | 1.96 A X-ray structure of photosynthetic reaction center from Rhodopseudomonas viridis:Crystals grown by microfluidic technique | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Li, L, Mustafi, D, Fu, Q, Tereshko, V, Chen, D.L, Tice, J.D, Ismagilov, R.F. | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Nanoliter microfluidic hybrid method for simultaneous screening and optimization validated with crystallization of membrane proteins.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I2H
 
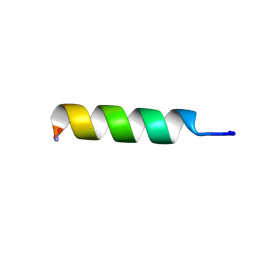 | | NMR structure of TPC3 in TFE | | Descriptor: | signaling peptide TCP3 | | Authors: | Syvitski, R.T, Jakeman, D.L, Li, Y. | | Deposit date: | 2006-08-16 | | Release date: | 2006-10-17 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Analysis of Quorum-Sensing Signaling Peptides from Streptococcus mutans.
J.Bacteriol., 189, 2007
|
|
3OJ8
 
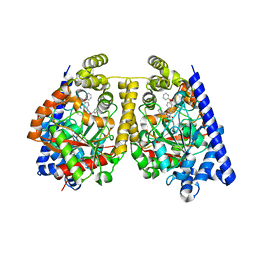 | | Alpha-Ketoheterocycle Inhibitors of Fatty Acid Amide Hydrolase Containing Additional Conformational Contraints in the Acyl Side Chain | | Descriptor: | (S)-[(2S)-6-phenoxy-1,2,3,4-tetrahydronaphthalen-2-yl](5-pyridin-2-yl-1,3-oxazol-2-yl)methanol, CHLORIDE ION, Fatty-acid amide hydrolase 1 | | Authors: | Mileni, M, Stevens, R.C, Boger, D.L. | | Deposit date: | 2010-08-20 | | Release date: | 2011-07-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | alpha-Ketoheterocycle Inhibitors of Fatty Acid Amide Hydrolase Containing Additional Conformational Contraints in the Acyl Side Chain
J.Med.Chem., 54, 2011
|
|
3OQE
 
 | | Structure of OpdA mutant Y257F | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, Phosphotriesterase | | Authors: | Ely, F, Guddat, L.W, Ollis, D.L, Schenk, G. | | Deposit date: | 2010-09-02 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The organophosphate-degrading enzyme from Agrobacterium radiobacter displays mechanistic flexibility for catalysis.
Biochem.J., 432, 2010
|
|
3OX5
 
 | |
3ONA
 
 | | The SECRET domain in complex with CX3CL1 | | Descriptor: | CX3CL1 protein, Tumour necrosis factor receptor | | Authors: | Wang, X.Q, Xue, X.G, Wang, D.L. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of chemokine sequestration by CrmD, a poxvirus-encoded tumor necrosis factor receptor
Plos Pathog., 7, 2011
|
|
3OOD
 
 | | Structure of OpdA Y257F mutant soaked with diethyl 4-methoxyphenyl phosphate for 20 hours. | | Descriptor: | COBALT (II) ION, DIETHYL 4-METHOXYPHENYL PHOSPHATE, Phosphotriesterase | | Authors: | Ely, F, Guddat, L.W, Ollis, D.L, Schenk, G. | | Deposit date: | 2010-08-31 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The organophosphate-degrading enzyme from Agrobacterium radiobacter displays mechanistic flexibility for catalysis.
Biochem.J., 432, 2010
|
|
3OXQ
 
 | | Crystal Structure of Ca2+/CaM-CaV1.2 pre-IQ/IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin, Voltage-dependent L-type calcium channel subunit alpha-1C | | Authors: | Kim, E.Y, Rumpf, C.H, Van Petegem, F, Arant, R, Findeisen, F, Cooley, E.S, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2010-09-21 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Multiple C-terminal tail Ca(2+)/CaMs regulate Ca(V)1.2 function but do not mediate channel dimerization.
Embo J., 29, 2010
|
|
