2BSW
 
 | | Crystal structure of a glyphosate-N-acetyltransferase obtained by DNA shuffling. | | Descriptor: | GLYCEROL, GLYPHOSATE N-ACETYLTRANSFERASE, OXIDIZED COENZYME A, ... | | Authors: | Keenan, R.J, Siehl, D.L, Gorton, R, Castle, L.A. | | Deposit date: | 2005-05-24 | | Release date: | 2005-06-08 | | Last modified: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | DNA Shuffling as a Tool for Protein Crystallization.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5V2Q
 
 | | CaV beta2a subunit: CaV1.2 AID-CEN complex | | Descriptor: | 1,3-bis(bromomethyl)benzene, CHLORIDE ION, Voltage-dependent L-type calcium channel subunit alpha-1C, ... | | Authors: | Findeisen, F, Campiglio, M, Jo, H, Rumpf, C.H, Pope, L, Flucher, B, Degrado, W.F, Minor, D.L. | | Deposit date: | 2017-03-06 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Stapled Voltage-Gated Calcium Channel (CaV) alpha-Interaction Domain (AID) Peptides Act As Selective Protein-Protein Interaction Inhibitors of CaV Function.
ACS Chem Neurosci, 8, 2017
|
|
5VRF
 
 | | CryoEM Structure of the Zinc Transporter YiiP from helical crystals | | Descriptor: | Cadmium and zinc efflux pump FieF, ZINC ION | | Authors: | Coudray, N, Lopez-Redondo, M, Zhang, Z, Alexopoulos, J, Stokes, D.L, Transcontinental EM Initiative for Membrane Protein Structure (TEMIMPS) | | Deposit date: | 2017-05-10 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for the alternating access mechanism of the cation diffusion facilitator YiiP.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2D2J
 
 | | OpdA from Agrobacterium radiobacter without inhibitor/product present at 1.75 A resolution | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, phosphotriesterase | | Authors: | Jackson, C, Kim, H.K, Carr, P.D, Liu, J.W, Ollis, D.L. | | Deposit date: | 2005-09-09 | | Release date: | 2005-09-20 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of an enzyme-product complex reveals the critical role of a terminal hydroxide nucleophile in the bacterial phosphotriesterase mechanism
Biochim.Biophys.Acta, 1752, 2005
|
|
2D2G
 
 | | OpdA from Agrobacterium radiobacter with bound product dimethylthiophosphate | | Descriptor: | COBALT (II) ION, O,O-DIMETHYL HYDROGEN THIOPHOSPHATE, phosphotriesterase | | Authors: | Jackson, C, Kim, H.K, Carr, P.D, Liu, J.W, Ollis, D.L. | | Deposit date: | 2005-09-08 | | Release date: | 2005-09-20 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of an enzyme-product complex reveals the critical role of a terminal hydroxide nucleophile in the bacterial phosphotriesterase mechanism
Biochim.Biophys.Acta, 1752, 2005
|
|
5HK6
 
 | |
5HKD
 
 | | Bacterial sodium channel neck 7G mutant | | Descriptor: | CALCIUM ION, Ion transport protein | | Authors: | Rohaim, A, Minor, D.L. | | Deposit date: | 2016-01-13 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Unfolding of a Temperature-Sensitive Domain Controls Voltage-Gated Channel Activation.
Cell, 164, 2016
|
|
2D2H
 
 | | OpdA from Agrobacterium radiobacter with bound inhibitor trimethyl phosphate at 1.8 A resolution | | Descriptor: | COBALT (II) ION, TRIMETHYL PHOSPHATE, phosphotriesterase | | Authors: | Jackson, C, Kim, H.K, Carr, P.D, Liu, J.W, Ollis, D.L. | | Deposit date: | 2005-09-09 | | Release date: | 2005-09-20 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of an enzyme-product complex reveals the critical role of a terminal hydroxide nucleophile in the bacterial phosphotriesterase mechanism
Biochim.Biophys.Acta, 1752, 2005
|
|
5V2P
 
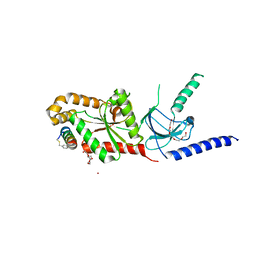 | | CaV beta2a subunit: CaV1.2 AID-CAP complex | | Descriptor: | 1,3-bis(bromomethyl)benzene, NICKEL (II) ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Findeisen, F, Campiglio, M, Jo, H, Rumpf, C.H, Pope, L, Flucher, B, Degrado, W.F, Minor, D.L. | | Deposit date: | 2017-03-06 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Stapled Voltage-Gated Calcium Channel (CaV) alpha-Interaction Domain (AID) Peptides Act As Selective Protein-Protein Interaction Inhibitors of CaV Function.
ACS Chem Neurosci, 8, 2017
|
|
5FUA
 
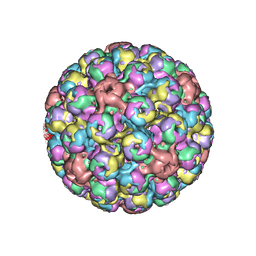 | | Cryo-EM of BK polyomavirus | | Descriptor: | MAJOR CAPSID PROTEIN VP1 | | Authors: | Hurdiss, D.L, Morgan, E.L, Thompson, R.F, Prescott, E.L, Panou, M.M, Macdonald, A, Ranson, N.A. | | Deposit date: | 2016-01-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | New Structural Insights Into the Genome and Minor Capsid Proteins of Bk Polyomavirus Using Cryo-Electron Microscopy.
Structure, 24, 2016
|
|
5HK7
 
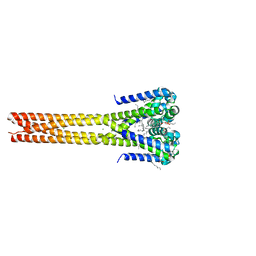 | | Bacterial sodium channel pore, 2.95 Angstrom resolution | | Descriptor: | 2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}ethyl heptadecanoate, CHLORIDE ION, Ion transport protein, ... | | Authors: | Shaya, D, Findeisen, F, Rohaim, A, Minor, D.L. | | Deposit date: | 2016-01-14 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Unfolding of a Temperature-Sensitive Domain Controls Voltage-Gated Channel Activation.
Cell, 164, 2016
|
|
2BKE
 
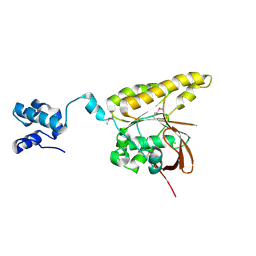 | | Conformational Flexibility Revealed by the Crystal Structure of a Crenarchaeal RadA | | Descriptor: | CHLORIDE ION, DNA REPAIR AND RECOMBINATION PROTEIN RADA | | Authors: | Ariza, A, Richard, D.L, White, M.F, Bond, C.S. | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational Flexibility Revealed by the Crystal Structure of a Crenarchaeal Rada
Nucleic Acids Res., 33, 2005
|
|
5W97
 
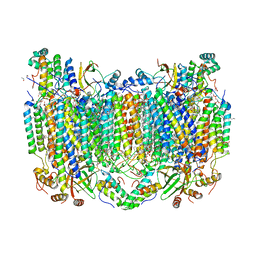 | | Crystal Structure of CO-bound Cytochrome c Oxidase determined by Serial Femtosecond X-Ray Crystallography at Room Temperature | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Rousseau, D.L, Yeh, S.-R, Ishigami, I, Zatsepin, N.A, Grant, T.D, Fromme, P, Fromme, R. | | Deposit date: | 2017-06-22 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CO-bound cytochrome c oxidase determined by serial femtosecond X-ray crystallography at room temperature.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VHU
 
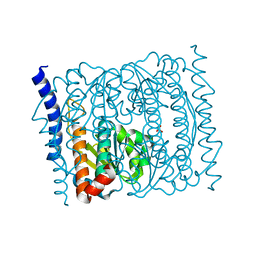 | | E. coli CFT073 c3406 | | Descriptor: | Isomerase, SULFATE ION | | Authors: | Cech, D.L, Pratt, A.C, Woodard, R.W. | | Deposit date: | 2017-04-13 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the mechanism of arabinose-5-phosphate isomerases.
To Be Published
|
|
5HJ8
 
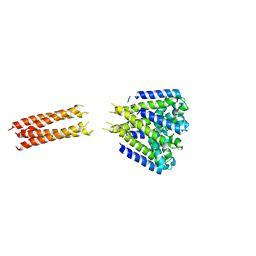 | | Bacterial sodium channel neck 3G mutant | | Descriptor: | CALCIUM ION, Ion transport protein | | Authors: | Rohaim, A, Minor, D.L. | | Deposit date: | 2016-01-13 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Unfolding of a Temperature-Sensitive Domain Controls Voltage-Gated Channel Activation.
Cell, 164, 2016
|
|
5WBF
 
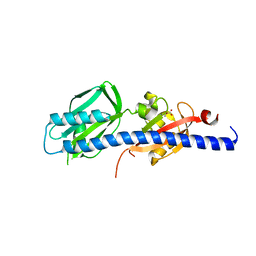 | | Double CACHE (dCACHE) sensing domain of TlpC chemoreceptor from Helicobacter pylori | | Descriptor: | GLYCEROL, LACTIC ACID, Methyl-accepting chemotaxis transducer (TlpC) | | Authors: | Machuca, M.A, Johnson, K.S, Liu, Y.C, Steer, D.L, Ottemann, K.M, Roujeinikova, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Helicobacter pylori chemoreceptor TlpC mediates chemotaxis to lactate.
Sci Rep, 7, 2017
|
|
5WUD
 
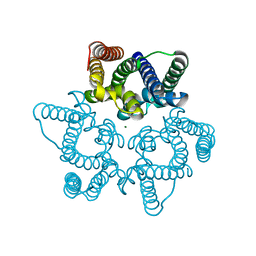 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
2ADB
 
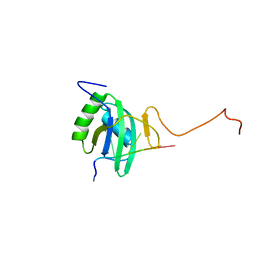 | | Solution structure of Polypyrimidine Tract Binding protein RBD2 complexed with CUCUCU RNA | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*U)-3', Polypyrimidine tract-binding protein 1 | | Authors: | Oberstrass, F.C, Auweter, S.D, Erat, M, Hargous, Y, Henning, A, Wenter, P, Reymond, L, Pitsch, S, Black, D.L, Allain, F.H.T. | | Deposit date: | 2005-07-20 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of PTB bound to RNA: specific binding and implications for splicing regulation
Science, 309, 2005
|
|
5WUE
 
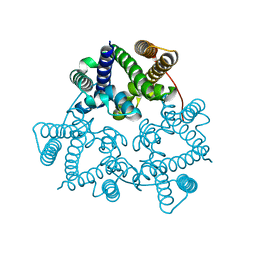 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
5IJC
 
 | | The crystal structure of mouse TLR4/MD-2/neoseptin-3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2BE6
 
 | | 2.0 A crystal structure of the CaV1.2 IQ domain-Ca/CaM complex | | Descriptor: | CALCIUM ION, Calmodulin 2, NICKEL (II) ION, ... | | Authors: | Van Petegem, F, Chatelain, F.C, Minor Jr, D.L. | | Deposit date: | 2005-10-23 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into voltage-gated calcium channel regulation from the structure of the Ca(V)1.2 IQ domain-Ca(2+)/calmodulin complex
Nat.Struct.Mol.Biol., 12, 2005
|
|
5IJB
 
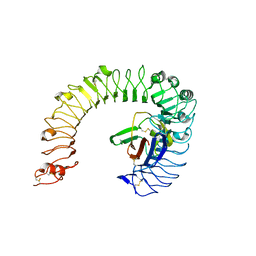 | | The ligand-free structure of the mouse TLR4/MD-2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5UQI
 
 | |
2A11
 
 | |
2ADC
 
 | | Solution structure of Polypyrimidine Tract Binding protein RBD34 complexed with CUCUCU RNA | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*U)-3', Polypyrimidine tract-binding protein 1 | | Authors: | Oberstrass, F.C, Auweter, S.D, Erat, M, Hargous, Y, Henning, A, Wenter, P, Reymond, L, Pitsch, S, Black, D.L, Allain, F.H.T. | | Deposit date: | 2005-07-20 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of PTB bound to RNA: specific binding and implications for splicing regulation
Science, 309, 2005
|
|
