4JNM
 
 | | Discovery of Potent and Efficacious Urea-containing Nicotinamide Phosphoribosyltransferase (NAMPT) Inhibitors with Reduced CYP2C9 Inhibition Properties | | Descriptor: | 1,2-ETHANEDIOL, 1-[(6-aminopyridin-3-yl)methyl]-3-[4-(phenylsulfonyl)phenyl]urea, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Gunzner-Toste, J, Zhao, G, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Fu, B, Han, B, Ho, Y, Kley, N, Liederer, B, Lin, J, Mukadam, S, O'Brien, T, Reynolds, D.J, Sharma, G, Skelton, N, Smith, C.C, Oh, A, Wang, W, Wang, Z, Xiao, Y, Yuen, P, Zak, M, Zhang, L, Zheng, X, Bair, K.W, Dragovich, P.S. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of potent and efficacious urea-containing nicotinamide phosphoribosyltransferase (NAMPT) inhibitors with reduced CYP2C9 inhibition properties.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4JS0
 
 | | Complex of Cdc42 with the CRIB-PR domain of IRSp53 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Brain-specific angiogenesis inhibitor 1-associated protein 2, Cell division control protein 42 homolog, ... | | Authors: | Kast, D.J, Dominguez, R. | | Deposit date: | 2013-03-22 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of IRSp53 inhibition and combinatorial activation by Cdc42 and downstream effectors.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4KSC
 
 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KQP
 
 | | Crystal structure of Lactococcus lactis GlnP substrate binding domain 2 (SBD2) in complex with glutamine at 0.95 A resolution | | Descriptor: | GLUTAMINE, Glutamine ABC transporter permease and substrate binding protein protein | | Authors: | Vujicic Zagar, A, Guskov, A, Schuurman-Wolters, G.K, Slotboom, D.J, Poolman, B. | | Deposit date: | 2013-05-15 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Functional Diversity of Tandem Substrate-Binding Domains in ABC Transporters from Pathogenic Bacteria.
Structure, 21, 2013
|
|
4KSB
 
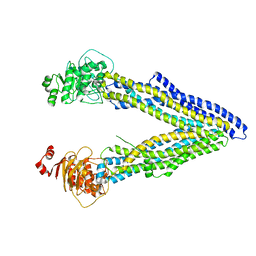 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8001 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KNS
 
 | | Reduced crystal structure of the Nitrosomonas europaea copper nitrite reductase at pH 6.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Rosenzweig, A.C, Lawton, T.L, Sayavedra-Soto, L.A, Arp, D.J. | | Deposit date: | 2013-05-10 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of a nitrite reductase involved in nitrifier denitrification.
J.Biol.Chem., 288, 2013
|
|
4K9B
 
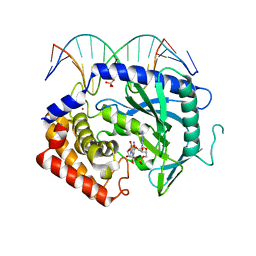 | | Structure of Ternary Complex of cGAS with dsDNA and Bound c[G(2 ,5 )pA(3 ,5 )p] | | Descriptor: | ADENOSINE MONOPHOSPHATE, Cyclic GMP-AMP synthase, DNA-F, ... | | Authors: | Gao, P, Wu, Y, Patel, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase.
Cell(Cambridge,Mass.), 153, 2013
|
|
4KPT
 
 | | Crystal structure of substrate binding domain 1 (SBD1) OF ABC transporter GLNPQ from lactococcus lactis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vujicic Zagar, A, Guskov, A, Schuurman-Wolters, G.K, Slotboom, D.J, Poolman, B. | | Deposit date: | 2013-05-14 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Functional Diversity of Tandem Substrate-Binding Domains in ABC Transporters from Pathogenic Bacteria.
Structure, 21, 2013
|
|
4KR5
 
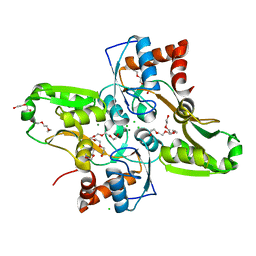 | | Crystal structure of Lactococcus lactis GlnP substrate binding domain 2 (SBD2) in open conformation | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Vujicic Zagar, A, Guskov, A, Schuurman-Wolters, G.K, Slotboom, D.J, Poolman, B. | | Deposit date: | 2013-05-16 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional Diversity of Tandem Substrate-Binding Domains in ABC Transporters from Pathogenic Bacteria.
Structure, 21, 2013
|
|
4KBS
 
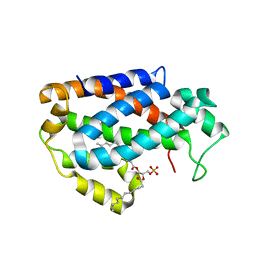 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 12:0 phosphatidic acid (12:0 PA) | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-ETHANEDIOL, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4K96
 
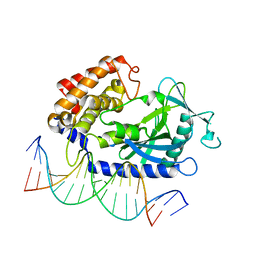 | | Structure of Binary Complex of cGAS with Bound dsDNA | | Descriptor: | Cyclic GMP-AMP synthase, DNA-F, DNA-R, ... | | Authors: | Gao, P, Wu, Y, Patel, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase.
Cell(Cambridge,Mass.), 153, 2013
|
|
4KNT
 
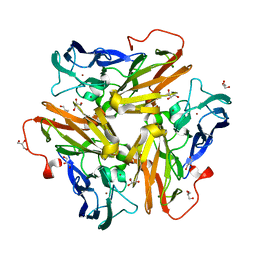 | | Copper nitrite reductase from Nitrosomonas europaea pH 8.5 | | Descriptor: | COPPER (II) ION, GLYCEROL, Multicopper oxidase type 1 | | Authors: | Rosenzweig, A.C, Lawton, T.L, Sayavedra-Soto, L.A, Arp, D.J. | | Deposit date: | 2013-05-10 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a nitrite reductase involved in nitrifier denitrification.
J.Biol.Chem., 288, 2013
|
|
4KNU
 
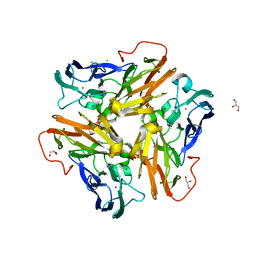 | | Copper nitrite reductase from Nitrosomonas europaea at pH 6.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Rosenzweig, A.C, Lawton, T.L, Sayavedra-Soto, L.A, Arp, D.J. | | Deposit date: | 2013-05-10 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a nitrite reductase involved in nitrifier denitrification.
J.Biol.Chem., 288, 2013
|
|
4K98
 
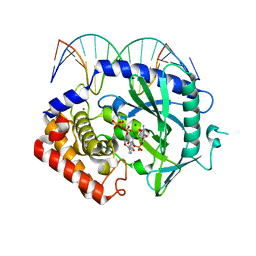 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppG(2 ,5 )pG | | Descriptor: | Cyclic GMP-AMP synthase, DNA-F, DNA-R, ... | | Authors: | Gao, P, Wu, Y, Patel, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase.
Cell(Cambridge,Mass.), 153, 2013
|
|
4KF6
 
 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 8:0 Ceramide-1-Phosphate (8:0-C1P) | | Descriptor: | (2S,3R,4E)-3-hydroxy-2-(octanoylamino)octadec-4-en-1-yl dihydrogen phosphate, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-26 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.195 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
4KFN
 
 | | Structure-Based Discovery of Novel Amide-Containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-[4-(piperidin-1-ylsulfonyl)benzyl]-1H-pyrrolo[3,2-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Gunzner-Toste, J, Liederer, B.M, Ly, J, O'Brien, T, Oh, A, Wang, L, Wang, W, Xiao, Y, Zak, M, Zhao, G, Yuen, P, Bair, K.W. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
4LBE
 
 | | Structure of KcsA with R122A mutation | | Descriptor: | DIACYL GLYCEROL, Fab heavy chain, Fab light chain, ... | | Authors: | Nimigean, C.M, Posson, D.J, McCoy, J.M. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-30 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Molecular interactions involved in proton-dependent gating in KcsA potassium channels.
J.Gen.Physiol., 142, 2013
|
|
4LA9
 
 | | Crystal structure of an empty substrate binding domain 1 (SBD1) OF ABC TRANSPORTER GLNPQ from lactococcus lactis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glutamine ABC transporter permease and substrate binding protein protein, PHOSPHATE ION | | Authors: | Guskov, A, Schuurman-Wolters, G.K, Slotboom, D.J, Poolman, B. | | Deposit date: | 2013-06-19 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Functional Diversity of Tandem Substrate-Binding Domains in ABC Transporters from Pathogenic Bacteria.
Structure, 21, 2013
|
|
4KXT
 
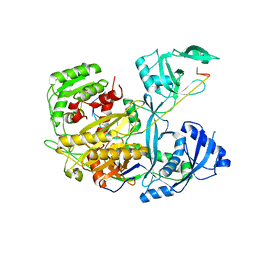 | |
4KY0
 
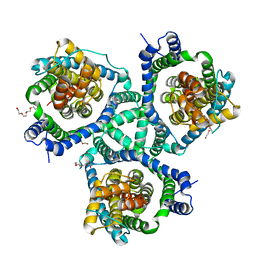 | | Crystal structure of a substrate-free glutamate transporter homologue from Thermococcus kodakarensis | | Descriptor: | Proton/glutamate symporter, SDF family, TETRAETHYLENE GLYCOL | | Authors: | Guskov, A, Jensen, S, Rempel, S, Hanelt, I, Slotboom, D.J. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a substrate-free aspartate transporter.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4LCU
 
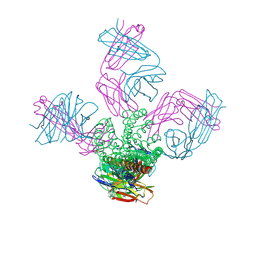 | | Structure of KcsA with E118A mutation | | Descriptor: | DIACYL GLYCEROL, Fab heavy chain, Fab light chain, ... | | Authors: | Nimigean, C.M, Posson, D.J, McCoy, J.M. | | Deposit date: | 2013-06-23 | | Release date: | 2013-10-30 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | Molecular interactions involved in proton-dependent gating in KcsA potassium channels.
J.Gen.Physiol., 142, 2013
|
|
4LOH
 
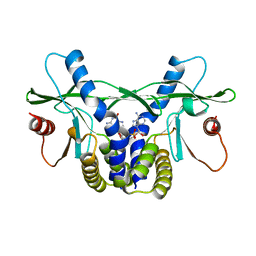 | | Crystal structure of hSTING(H232) in complex with c[G(2',5')pA(3',5')p] | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
4LOJ
 
 | | Crystal structure of mSting in complex with c[G(2',5')pA(3',5')p] | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
4JOL
 
 | |
4MDT
 
 | | Structure of LpxC bound to the reaction product UDP-(3-O-(R-3-hydroxymyristoyl))-glucosamine | | Descriptor: | PHOSPHATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION, ... | | Authors: | Clayton, G.M, Klein, D.J, Rickert, K.W, Patel, S.B, Kornienko, M, Zugay-Murphy, J, Reid, J.C, Tummala, S, Sharma, S, Singh, S.B, Miesel, L, Lumb, K.J, Soisson, S.M. | | Deposit date: | 2013-08-23 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of the Bacterial Deacetylase LpxC Bound to the Nucleotide Reaction Product Reveals Mechanisms of Oxyanion Stabilization and Proton Transfer.
J.Biol.Chem., 288, 2013
|
|
