3GK4
 
 | | X-ray structure of bovine SBi523,Ca(2+)-S100B | | Descriptor: | CALCIUM ION, Protein S100-B, ethyl 5-{[(1R)-1-(ethoxycarbonyl)-2-oxopropyl]sulfanyl}-1,2-dihydro[1,2,3]triazolo[1,5-a]quinazoline-3-carboxylate | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-03-09 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
Biochemistry, 48, 2009
|
|
3GK1
 
 | | X-ray structure of bovine SBi132,Ca(2+)-S100B | | Descriptor: | 2-[(5-hex-1-yn-1-ylfuran-2-yl)carbonyl]-N-methylhydrazinecarbothioamide, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-03-09 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small molecules bound to unique sites in the target protein binding cleft of calcium-bound S100B as characterized by nuclear magnetic resonance and X-ray crystallography.
Biochemistry, 48, 2009
|
|
3UP0
 
 | | Nuclear receptor DAF-12 from hookworm Ancylostoma ceylanicum in complex with (25S)-delta7-dafachronic acid | | Descriptor: | (5beta,14beta,17alpha,25S)-3-oxocholest-7-en-26-oic acid, Nuclear receptor coactivator 2, aceDAF-12 | | Authors: | Zhi, X, Zhou, X.E, Melcher, K, Motola, D.L, Gelmedin, V, Hawdon, J, Kliewer, S.A, Mangelsdorf, D.J, Xu, H.E. | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Conservation of Ligand Binding Reveals a Bile Acid-like Signaling Pathway in Nematodes.
J.Biol.Chem., 287, 2012
|
|
3GWX
 
 | | MOLECULAR RECOGNITION OF FATTY ACIDS BY PEROXISOME PROLIFERATOR-ACTIVATED RECEPTORS | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, PROTEIN (PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR (PPAR-DELTA)) | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Parks, D.J, Blanchard, S.G, Brown, P.J, Sternbach, D.D, Lehmann, J.M, Wisely, G.B, Willson, T.M, Kliewer, S.A, Milburn, M.V. | | Deposit date: | 1999-04-26 | | Release date: | 2000-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition of fatty acids by peroxisome proliferator-activated receptors.
Mol.Cell, 3, 1999
|
|
3VBH
 
 | | Crystal structure of formaldehyde treated human enterovirus 71 (space group R32) | | Descriptor: | CHLORIDE ION, Genome Polyprotein, capsid protein VP1, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3U2P
 
 | | Crystal structure of N-terminal three extracellular domains of ErbB4/Her4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor tyrosine-protein kinase erbB-4 | | Authors: | Liu, P, Bouyain, S, Elgenbrot, C, Leahy, D.J. | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The ErbB4 extracellular region retains a tethered-like conformation in the absence of the tether.
Protein Sci., 21, 2012
|
|
3IQQ
 
 | | X-ray structure of bovine TRTK12-Ca(2+)-S100B | | Descriptor: | CALCIUM ION, Protein S100-B, TRTK12 peptide, ... | | Authors: | Charpentier, T.H, Weber, D.J, Toth, E.A. | | Deposit date: | 2009-08-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Effects of CapZ Peptide (TRTK-12) Binding to S100B-Ca(2+) as Examined by NMR and X-ray Crystallography
J.Mol.Biol., 396, 2010
|
|
3TCH
 
 | | Crystal structure of E. coli OppA in an open conformation | | Descriptor: | Periplasmic oligopeptide-binding protein | | Authors: | Klepsch, M.M, Kovermann, M, Low, C, Balbach, J, de Gier, J.W, Slotboom, D.J, Berntsson, R.P.-A. | | Deposit date: | 2011-08-09 | | Release date: | 2011-10-12 | | Last modified: | 2012-01-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Escherichia coli peptide binding protein OppA has a preference for positively charged peptides.
J.Mol.Biol., 414, 2011
|
|
3SOX
 
 | | Structure of UHRF1 PHD finger in the free form | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Rajakumara, E, Patel, D.J. | | Deposit date: | 2011-06-30 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6501 Å) | | Cite: | PHD Finger Recognition of Unmodified Histone H3R2 Links UHRF1 to Regulation of Euchromatic Gene Expression.
Mol.Cell, 43, 2011
|
|
3IEP
 
 | | Firefly luciferase apo structure (P41 form) | | Descriptor: | Luciferin 4-monooxygenase | | Authors: | Lovell, S, Battaile, K.P, Auld, D.S, Thorne, N, Lea, W.A, Maloney, D.J, Shen, M, Raj, G, Thomas, C.J, Simeonov, A, Hanzlik, R.P, Inglese, J. | | Deposit date: | 2009-07-23 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3HJF
 
 | | Crystal structure of T. thermophilus Argonaute E546 mutant protein complexed with DNA guide strand and 15-nt RNA target strand | | Descriptor: | 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', 5'-R(*CP*AP*AP*CP*CP*UP*AP*CP*UP*AP*CP*CP*UP*CP*G)-3', Argonaute, ... | | Authors: | Wang, Y, Li, H, Sheng, G, Patel, D.J. | | Deposit date: | 2009-05-21 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.056 Å) | | Cite: | Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes.
Nature, 461, 2009
|
|
3HF3
 
 | | Old Yellow Enzyme from Thermus scotoductus SA-01 | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE, SULFATE ION | | Authors: | Opperman, D.J, Sewell, B.T, Litthauer, D, Isupov, M.N, Littlechild, J.A, van Heerden, E. | | Deposit date: | 2009-05-11 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a thermostable old yellow enzyme from Thermus scotoductus SA-01
Biochem.Biophys.Res.Commun., 393, 2010
|
|
3TOS
 
 | | Crystal Structure of CalS11, Calicheamicin Methyltransferase | | Descriptor: | 1,2-ETHANEDIOL, CalS11, GLUTAMIC ACID, ... | | Authors: | Chang, A, Aceti, D.J, Beebe, E.T, Makino, S.-I, Wrobel, R.L, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-09-06 | | Release date: | 2011-10-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of CalS11, Calicheamicin methyltransferase
To be Published
|
|
3FC6
 
 | | hRXRalpha & mLXRalpha with an indole Pharmacophore, SB786875 | | Descriptor: | Nr1h3 protein, RETINOIC ACID, Retinoic acid receptor RXR-alpha, ... | | Authors: | Washburn, D.G, Hoang, T.H, Campobasso, N, Smallwood, A, Parks, D.J, Webb, C.L, Frank, K, Nord, M, Duraiswami, C, Evans, C, Jaye, M, Thompson, S.K. | | Deposit date: | 2008-11-21 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.063 Å) | | Cite: | Synthesis and SAR of potent LXR agonists containing an indole pharmacophore.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3IQO
 
 | |
3IVV
 
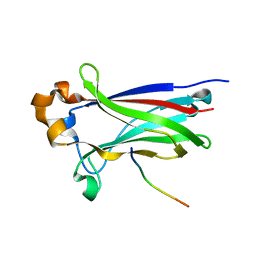 | | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATH-PucSBC1_pep1 | | Descriptor: | PucSBC1, Speckle-type POZ protein | | Authors: | Schulman, B.A, Miller, D.J, Calabrese, M.F, Seyedin, S. | | Deposit date: | 2009-09-01 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases.
Mol.Cell, 36, 2009
|
|
3UIJ
 
 | |
3UIH
 
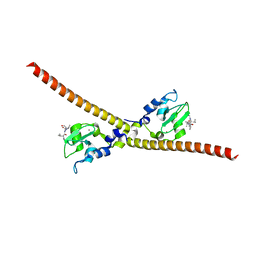 | |
3IVB
 
 | | Structures of SPOP-Substrate Complexes: Insights into Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATH-MacroH2ASBCpep1 | | Descriptor: | CHLORIDE ION, Core histone macro-H2A.1, Speckle-type POZ protein, ... | | Authors: | Schulman, B.A, Miller, D.J, Calabrese, M.F, Seyedin, S. | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure 9
To be Published
|
|
3IYC
 
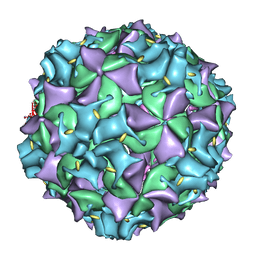 | | Poliovirus late RNA-release intermediate | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Genome polyprotein, ... | | Authors: | Levy, H.C, Bostina, M, Filman, D.J, Hogle, J.M. | | Deposit date: | 2009-07-21 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy.
J.Virol., 84, 2010
|
|
3K8E
 
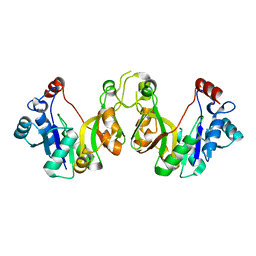 | | Crystal structure of E. coli lipopolysaccharide specific CMP-KDO synthetase | | Descriptor: | 3-deoxy-manno-octulosonate cytidylyltransferase | | Authors: | Heyes, D.J, Levy, C.W, Lafite, P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-10-14 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure-based mechanism of CMP-2-keto-3-deoxymanno-octulonic acid synthetase: convergent evolution of a sugar-activating enzyme with DNA/RNA polymerases
J.Biol.Chem., 284, 2009
|
|
3J41
 
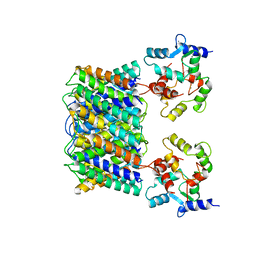 | | Pseudo-atomic model of the Aquaporin-0/Calmodulin complex derived from electron microscopy | | Descriptor: | CALCIUM ION, Calmodulin, Lens fiber major intrinsic protein | | Authors: | Reichow, S.L, Clemens, D.M, Freites, J.A, Nemeth-Cahalan, K.L, Heyden, M, Tobias, D.J, Hall, J.E, Gonen, T. | | Deposit date: | 2013-05-31 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Allosteric mechanism of water-channel gating by Ca(2+)-calmodulin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3VBS
 
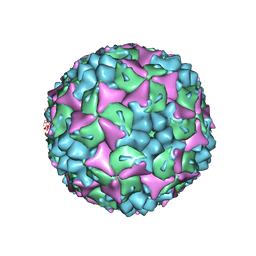 | | Crystal structure of human Enterovirus 71 | | Descriptor: | Genome Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2CWO
 
 | |
2ORX
 
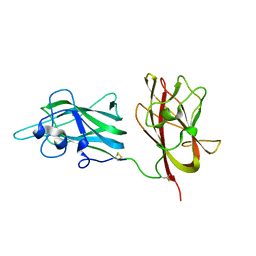 | | Structural Basis for Ligand Binding and Heparin Mediated Activation of Neuropilin | | Descriptor: | Neuropilin-1 | | Authors: | Vander Kooi, C.W, Jusino, M.A, Perman, B, Neau, D.B, Bellamy, H.D, Leahy, D.J. | | Deposit date: | 2007-02-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for ligand and heparin binding to neuropilin B domains
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
