2GWP
 
 | | High-resolution solution structure of the salt-bridge defficient mouse defensin (E15D)-Cryptdin4 | | Descriptor: | Defensin-related cryptdin 4 | | Authors: | Rosengren, K.J, Craik, D.J, Vogel, H.J, Daly, N.L, Ouellette, A.J. | | Deposit date: | 2006-05-05 | | Release date: | 2006-07-25 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of the conserved salt bridge in mammalian paneth cell alpha-defensins: solution structures of mouse CRYPTDIN-4 and (E15D)-CRYPTDIN-4.
J.Biol.Chem., 281, 2006
|
|
2H0Z
 
 | |
2HM1
 
 | | Crystal Structure of human beta-secretase (BACE) in the presence of an inhibitor (2) | | Descriptor: | Beta-secretase 1, N-{(1S)-2-({(1S,2R)-1-(3,5-DIFLUOROBENZYL)-3-[(3-ETHYLBENZYL)AMINO]-2-HYDROXYPROPYL}AMINO)-2-OXO-1-[(PENTYLSULFONYL)METHYL]ETHYL}NICOTINAMIDE | | Authors: | Benson, T.E, Prince, D.B, Tomasselli, A.G, Emmons, T.L, Paddock, D.J. | | Deposit date: | 2006-07-10 | | Release date: | 2007-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of potent inhibitors of human beta-secretase. Part 2.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2HT7
 
 | | N8 neuraminidase in open complex with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, Neuraminidase | | Authors: | Russell, R.J, Haire, L.F, Stevens, D.J, Collins, P.J, Lin, Y.P, Blackburn, G.M, Hay, A.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2006-07-25 | | Release date: | 2006-09-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design.
Nature, 443, 2006
|
|
2HO6
 
 | |
2HMN
 
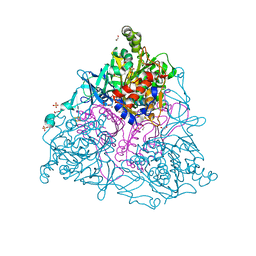 | | Crystal Structure of the Naphthalene 1,2-Dioxygenase F352V Mutant Bound to Anthracene. | | Descriptor: | 1,2-ETHANEDIOL, ANTHRACENE, FE (III) ION, ... | | Authors: | Ferraro, D.J, Okerlund, A.L, Mowers, J.C, Ramaswamy, S. | | Deposit date: | 2006-07-11 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for regioselectivity and stereoselectivity of product formation by naphthalene 1,2-dioxygenase.
J.Bacteriol., 188, 2006
|
|
2HTW
 
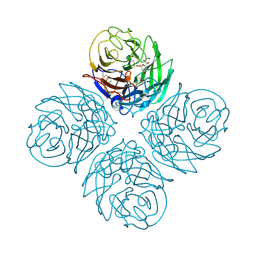 | | N4 neuraminidase in complex with DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, Neuraminidase | | Authors: | Russell, R.J, Haire, L.F, Stevens, D.J, Collins, P.J, Lin, Y.P, Blackburn, G.M, Hay, A.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design.
Nature, 443, 2006
|
|
2EVS
 
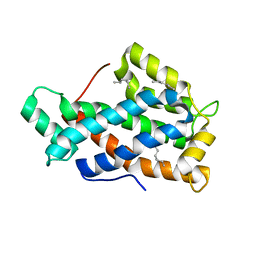 | | Crystal structure of human Glycolipid Transfer Protein complexed with n-hexyl-beta-D-glucoside | | Descriptor: | DECANE, Glycolipid transfer protein, HEXANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2F7T
 
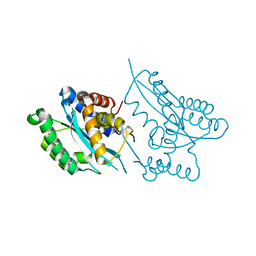 | | Crystal structure of the catalytic domain of Mos1 mariner transposase | | Descriptor: | MAGNESIUM ION, Mos1 transposase | | Authors: | Richardson, J.M, Dawson, A, Taylor, P, Finnegan, D.J, Walkinshaw, M.D. | | Deposit date: | 2005-12-01 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanism of Mos1 transposition: insights from structural analysis
Embo J., 25, 2006
|
|
2FHW
 
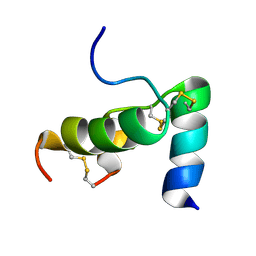 | | Solution structure of human relaxin-3 | | Descriptor: | Relaxin 3 (Prorelaxin H3) (Insulin-like peptide INSL7) (Insulin-like peptide 7) | | Authors: | Rosengren, K.J, Craik, D.J. | | Deposit date: | 2005-12-27 | | Release date: | 2006-01-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure and novel insights into the determinants of the receptor specificity of human relaxin-3.
J.Biol.Chem., 281, 2006
|
|
2F2J
 
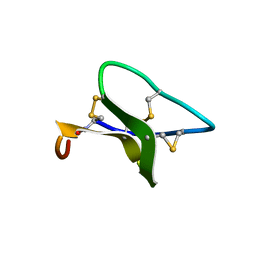 | |
2GK1
 
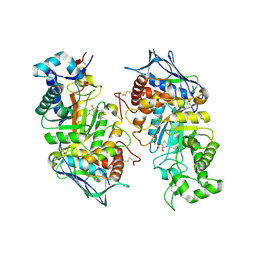 | | X-ray crystal structure of NGT-bound HexA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Beta-hexosaminidase subunit alpha, ... | | Authors: | Lemieux, M.J, Mark, B.L, Cherney, M.M, Withers, S.G, Mahuran, D.J, James, M.N. | | Deposit date: | 2006-03-31 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystallographic Structure of Human beta-Hexosaminidase A: Interpretation of Tay-Sachs Mutations and Loss of G(M2) Ganglioside Hydrolysis.
J.Mol.Biol., 359, 2006
|
|
2I1H
 
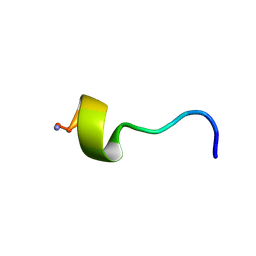 | | DPC micelle-bound NMR structures of Tritrp7 | | Descriptor: | 13-mer analogue of Prophenin-1 containing WWW | | Authors: | Schibli, D.J, Nguyen, L.T. | | Deposit date: | 2006-08-14 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of tritrpticin analogs: potential relationships between antimicrobial activities, model membrane interactions, and their micelle-bound NMR structures
Biophys.J., 91, 2006
|
|
2IBG
 
 | |
2IBB
 
 | |
2HU0
 
 | | N1 neuraminidase in complex with oseltamivir 1 | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, Neuraminidase | | Authors: | Russell, R.J, Haire, L.F, Stevens, D.J, Collins, P.J, Lin, Y.P, Blackburn, G.M, Hay, A.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design.
Nature, 443, 2006
|
|
2HTY
 
 | | N1 neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Russell, R.J, Haire, L.F, Stevens, D.J, Collins, P.J, Lin, Y.P, Blackburn, G.M, Hay, A.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design.
Nature, 443, 2006
|
|
2HU4
 
 | | N1 neuraminidase in complex with oseltamivir 2 | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, Neuraminidase | | Authors: | Russell, R.J, Haire, L.F, Stevens, D.J, Collins, P.J, Lin, Y.P, Blackburn, G.M, Hay, A.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design.
Nature, 443, 2006
|
|
2I1D
 
 | | DPC micelle-bound NMR structures of Tritrp1 | | Descriptor: | 13-mer from Prophenin-1 containing WWW | | Authors: | Schibli, D.J, Nguyen, L.T. | | Deposit date: | 2006-08-14 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of tritrpticin analogs: potential relationships between antimicrobial activities, model membrane interactions, and their micelle-bound NMR structures
Biophys.J., 91, 2006
|
|
2I1F
 
 | | DPC micelle-bound NMR structures of Tritrp3 | | Descriptor: | 13-mer analogue of Prophenin-1 containing WWW | | Authors: | Schibli, D.J, Nguyen, L.T. | | Deposit date: | 2006-08-14 | | Release date: | 2006-11-28 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of tritrpticin analogs: potential relationships between antimicrobial activities, model membrane interactions, and their micelle-bound NMR structures
Biophys.J., 91, 2006
|
|
2I13
 
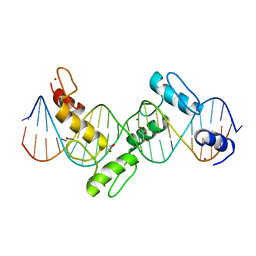 | | Aart, a six finger zinc finger designed to recognize ANN triplets | | Descriptor: | 5'-D(*CP*AP*GP*AP*TP*GP*TP*AP*GP*GP*GP*AP*AP*AP*AP*GP*CP*CP*CP*GP*GP*G)-3', 5'-D(*GP*CP*CP*CP*GP*GP*GP*CP*TP*TP*TP*TP*CP*CP*CP*TP*AP*CP*AP*TP*CP*T)-3', Aart, ... | | Authors: | Horton, N.C, Segal, D.J, Bhakta, M, Crotty, J.W, Barbas III, C.F. | | Deposit date: | 2006-08-12 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of Aart, a Designed Six-finger Zinc Finger Peptide, Bound to DNA.
J.Mol.Biol., 363, 2006
|
|
2I1G
 
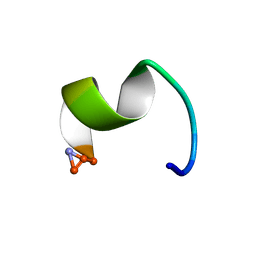 | | DPC micelle-bound NMR structures of Tritrp5 | | Descriptor: | 13-mer analogue of Prophenin-1 containing WWW | | Authors: | Schibli, D.J, Nguyen, L.T. | | Deposit date: | 2006-08-14 | | Release date: | 2006-11-28 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of tritrpticin analogs: potential relationships between antimicrobial activities, model membrane interactions, and their micelle-bound NMR structures
Biophys.J., 91, 2006
|
|
2I1I
 
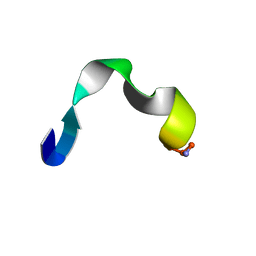 | | DPC micelle-bound NMR structures of Tritrp8 | | Descriptor: | 13-mer analogue of Prophenin-1 containing WWW | | Authors: | Schibli, D.J, Nguyen, L.T. | | Deposit date: | 2006-08-14 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of tritrpticin analogs: potential relationships between antimicrobial activities, model membrane interactions, and their micelle-bound NMR structures
Biophys.J., 91, 2006
|
|
2F2I
 
 | |
2F6N
 
 | |
