1BH4
 
 | |
2JZM
 
 | |
4FG2
 
 | |
4EYU
 
 | |
6CKS
 
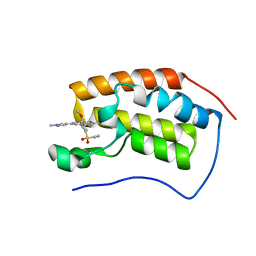 | | Crystal Structure of BRD4 with QC4956 | | Descriptor: | 4-[5-(ethylsulfonyl)-2-methoxyphenyl]-2-methyl-6-(1-methyl-1H-pyrazol-4-yl)isoquinolin-1(2H)-one, Bromodomain-containing protein 4 | | Authors: | Hosfield, D.J. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Design, synthesis and biological evaluation of novel 4-phenylisoquinolinone BET bromodomain inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
9BCC
 
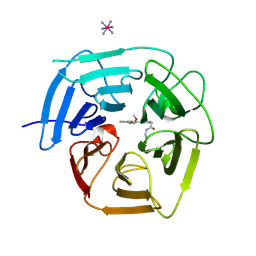 | | Structure of KLHDC2 bound to SJ46418 | | Descriptor: | COBALT HEXAMMINE(III), Kelch domain-containing protein 2, N-({2-[8-(2-methoxyethoxy)naphthalen-2-yl]-1,3-thiazol-4-yl}acetyl)glycine | | Authors: | Scott, D.C, Dharuman, S, Griffith, E, Chai, S.C, Ronnebaum, J, King, M.T, Tangallapally, R, Lee, C, Gee, C.T, Lee, H.W, Ochoada, J, Miller, D.J, Jayasinghe, T, Paulo, J.A, Elledge, S.J, Harper, J.W, Chen, T, Lee, R.E, Schulman, B.A. | | Deposit date: | 2024-04-08 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Principles of isoform-specific targeted protein degradation engaging the C-degron E3 KLHDC2
To Be Published
|
|
201D
 
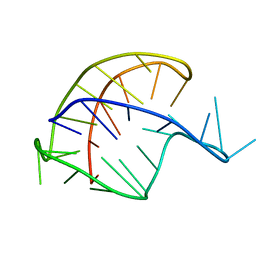 | |
202D
 
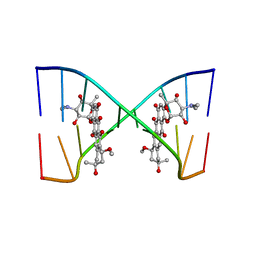 | |
1LKX
 
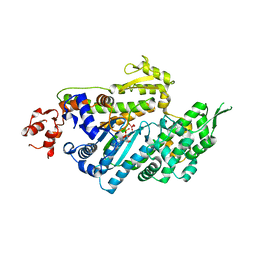 | | MOTOR DOMAIN OF MYOE, A CLASS-I MYOSIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN IE HEAVY CHAIN, ... | | Authors: | Kollmar, M, Durrwang, U, Kliche, W, Manstein, D.J, Kull, F.J. | | Deposit date: | 2002-04-26 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the motor domain of a class-I myosin.
EMBO J., 21, 2002
|
|
4FG4
 
 | |
6DGQ
 
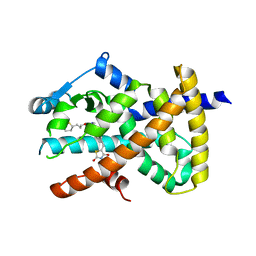 | | Crystal Structure of Human PPARgamma Ligand Binding Domain in Complex with CAY10506 | | Descriptor: | N-(2-{4-[(2,4-dioxo-3,4-dihydro-2H-1lambda~4~,3-thiazol-5-yl)methyl]phenoxy}ethyl)-5-[(3R)-1,2-dithiolan-3-yl]pentanamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Shang, J, Kojetin, D.J. | | Deposit date: | 2018-05-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Quantitative structural assessment of graded receptor agonism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6DKY
 
 | | Crystal structure of ribifolin, an orbitide from Jatropha ribifolia | | Descriptor: | ILE-LEU-GLY-SER-ILE-ILE-LEU-GLY | | Authors: | Wang, C.K, Ramalho, S.D, King, G.J, Craik, D.J. | | Deposit date: | 2018-05-31 | | Release date: | 2018-11-07 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.239 Å) | | Cite: | Synthesis, Racemic X-ray Crystallographic, and Permeability Studies of Bioactive Orbitides from Jatropha Species.
J. Nat. Prod., 81, 2018
|
|
1NL4
 
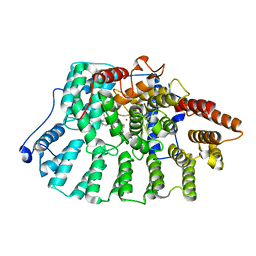 | | Crystal Structure of Rat Farnesyl Transferase in Complex With A Potent Biphenyl Inhibitor | | Descriptor: | 4-[(3-CYANO-BENZYL)-(3-METHYL-3H-IMIDAZOL-4-YLMETHYL)-AMINO]-2-NAPHTHALEN-1-YL-BENZONITRILE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, Protein farnesyltransferase alpha subunit, ... | | Authors: | Curtin, M.L, Florjancic, A.S, Cohen, J, Gu, W.-J, Frost, D.J, Muchmore, S.W, Sham, H.L. | | Deposit date: | 2003-01-06 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel and Selective Imidazole-containing Biphenyl Inhibitors of Protein Farnesyltransferase
BIOORG.MED.CHEM.LETT., 13, 2003
|
|
1AG7
 
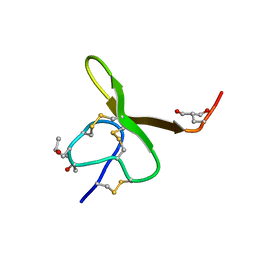 | | CONOTOXIN GS, NMR, 20 STRUCTURES | | Descriptor: | CONOTOXIN GS | | Authors: | Hill, J.M, Alewood, P.F, Craik, D.J. | | Deposit date: | 1997-04-03 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the sodium channel antagonist conotoxin GS: a new molecular caliper for probing sodium channel geometry.
Structure, 5, 1997
|
|
1NBJ
 
 | | High-resolution solution structure of cycloviolacin O1 | | Descriptor: | cycloviolacin O1 | | Authors: | Rosengren, K.J, Daly, N.L, Plan, M.R, Waine, C, Craik, D.J. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Twists, Knots, and Rings in Proteins. STRUCTURAL DEFINITION OF THE CYCLOTIDE FRAMEWORK.
J.Biol.Chem., 278, 2003
|
|
2BMR
 
 | | The Crystal Structure of Nitrobenzene Dioxygenase in complex with 3- nitrotoluene | | Descriptor: | 1,2-ETHANEDIOL, 3-NITROTOLUENE, ETHANOL, ... | | Authors: | Friemann, R, Ivkovic-Jensen, M.M, Lessner, D.J, Yu, C, Gibson, D.T, Parales, R.E, Eklund, H, Ramaswamy, S. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insight into the dioxygenation of nitroarene compounds: the crystal structure of nitrobenzene dioxygenase.
J. Mol. Biol., 348, 2005
|
|
2BRW
 
 | |
2BW0
 
 | | Crystal Structure of the hydrolase domain of Human 10-Formyltetrahydrofolate 2 dehydrogenase | | Descriptor: | 10-FORMYLTETRAHYDROFOLATE DEHYDROGENASE, SULFATE ION | | Authors: | Ogg, D.J, Stenmark, P, Arrowsmith, C, Edwards, A, Ehn, M, Graslund, S, Hammarstrom, M, Hallberg, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Persson, C, Sagemark, J, Schuler, H, Sundstrom, M, Thorsell, A, Dobritzsch, D, Weigelt, J. | | Deposit date: | 2005-07-07 | | Release date: | 2005-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the Hydrolase Domain of Human 10-Formyltetrahydrofolate Dehydrogenase and its Complex with a Substrate Analogue.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1NA8
 
 | | Crystal structure of ADP-ribosylation factor binding protein GGA1 | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Lui, W.W, Collins, B.M, Hirst, J, Motley, A, Millar, C, Schu, P, Owen, D.J, Robinson, M.S. | | Deposit date: | 2002-11-27 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding partners for the COOH-terminal appendage domains of the GGAs and gamma-adaptin
Mol.Cell.Biol., 14, 2003
|
|
6DHR
 
 | |
1NI6
 
 | | Comparisions of the Heme-Free and-Bound Crystal Structures of Human Heme Oxygenase-1 | | Descriptor: | CHLORIDE ION, Heme oxygenase 1, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Lad, L, Schuller, D.J, Friedman, J, Li, H, Shimizu, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2002-12-21 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comparison of the heme-free and -bound crystal structures of human heme oxygenase-1
J.Biol.Chem., 278, 2003
|
|
4GZ5
 
 | | Crystal structure of human O-GlcNAc Transferase with UDP-GlcNAc | | Descriptor: | SULFATE ION, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Lazarus, M.B, Jiang, J, Gloster, T.M, Zandberg, W.F, Vocadlo, D.J, Walker, S. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.075 Å) | | Cite: | Structural snapshots of the reaction coordinate for O-GlcNAc transferase.
Nat.Chem.Biol., 8, 2012
|
|
4GUX
 
 | | Crystal structure of trypsin:MCoTi-II complex | | Descriptor: | ACETATE ION, CALCIUM ION, Cationic trypsin, ... | | Authors: | King, G.J, Daly, N.L, Thorstholm, L, Greenwood, K.P, Rosengren, K.J, Heras, B, Craik, D.J, Martin, J.L. | | Deposit date: | 2012-08-30 | | Release date: | 2013-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural insights into the role of the cyclic backbone in a squash trypsin inhibitor
J.Biol.Chem., 288, 2013
|
|
1ZL9
 
 | | Crystal Structure of a major nematode C.elegans specific GST (CE01613) | | Descriptor: | GLUTATHIONE, glutathione S-transferase 5 | | Authors: | Kriksunov, I.A, Liu, Q, Schuller, D.J, Campbell, A.M, Barrett, J, Brophy, P.M, Hao, Q. | | Deposit date: | 2005-05-05 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of a major nematode C.elegans specific GST (CE01613)
To be Published
|
|
177D
 
 | |
