2PHK
 
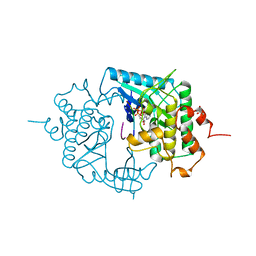 | | THE CRYSTAL STRUCTURE OF A PHOSPHORYLASE KINASE PEPTIDE SUBSTRATE COMPLEX: KINASE SUBSTRATE RECOGNITION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lowe, E.D, Noble, M.E.M, Skamnaki, V.T, Oikonomakos, N.G, Owen, D.J, Johnson, L.N. | | Deposit date: | 1998-06-18 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of a phosphorylase kinase peptide substrate complex: kinase substrate recognition.
EMBO J., 16, 1997
|
|
2PTX
 
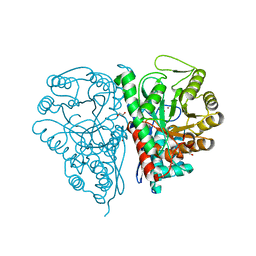 | | Crystal Structure of the T. brucei enolase complexed with sulphate in closed conformation | | Descriptor: | 1,2-ETHANEDIOL, Enolase, SULFATE ION, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
2PTZ
 
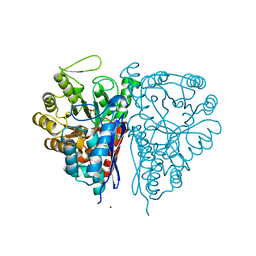 | | Crystal Structure of the T. brucei enolase complexed with phosphonoacetohydroxamate (PAH), His156-out conformation | | Descriptor: | 1,2-ETHANEDIOL, Enolase, PHOSPHONOACETOHYDROXAMIC ACID, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
2PTY
 
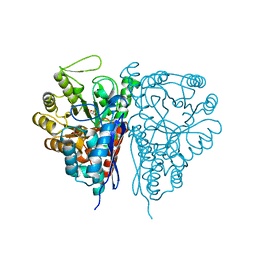 | | Crystal Structure of the T. brucei enolase complexed with PEP | | Descriptor: | 1,2-ETHANEDIOL, Enolase, PHOSPHOENOLPYRUVATE, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
2PU0
 
 | | Crystal Structure of the T. brucei enolase complexed with phosphonoacetohydroxamate (PAH), His156-in conformation | | Descriptor: | 1,2-ETHANEDIOL, Enolase, PHOSPHONOACETOHYDROXAMIC ACID, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
2PZL
 
 | | Crystal structure of the Bordetella bronchiseptica enzyme WbmG in complex with NAD and UDP | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative nucleotide sugar epimerase/ dehydratase, URIDINE-5'-DIPHOSPHATE | | Authors: | King, J.D, Harmer, N.J, Maskell, D.J, Blundell, T.L. | | Deposit date: | 2007-05-18 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Predicting protein function from structure--the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis.
J.Mol.Biol., 374, 2007
|
|
2PZK
 
 | | Crystal structure of the Bordetella bronchiseptica enzyme WbmG in complex with NAD | | Descriptor: | MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative nucleotide sugar epimerase/ dehydratase | | Authors: | King, J.D, Harmer, N.J, Maskell, D.J, Blundell, T.L. | | Deposit date: | 2007-05-18 | | Release date: | 2007-10-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Predicting protein function from structure--the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis.
J.Mol.Biol., 374, 2007
|
|
2PU1
 
 | | Crystal Structure of the T. brucei enolase complexed with Fluoro-phosphonoacetohydroxamate (FPAH) | | Descriptor: | 1,2-ETHANEDIOL, Enolase, ZINC ION, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
2PTA
 
 | | PANDINUS TOXIN K-A (PITX-KA) FROM PANDINUS IMPERATOR, NMR, 20 STRUCTURES | | Descriptor: | PANDINUS TOXIN K-ALPHA | | Authors: | Tenenholz, T.C, Rogowski, R.S, Collins, J.H, Blaustein, M.P, Weber, D.J. | | Deposit date: | 1996-11-26 | | Release date: | 1997-12-10 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure for Pandinus toxin K-alpha (PiTX-K alpha), a selective blocker of A-type potassium channels.
Biochemistry, 36, 1997
|
|
2GWX
 
 | | MOLECULAR RECOGNITION OF FATTY ACIDS BY PEROXISOME PROLIFERATOR-ACTIVATED RECEPTORS | | Descriptor: | PROTEIN (PPAR-DELTA) | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Park, D.J, Blanchard, S, Brown, P, Sternbach, D, Lehmann, J, Bruce, G.W, Willson, T.M, Kliewer, S.A, Milburn, M.V. | | Deposit date: | 1999-03-11 | | Release date: | 2000-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular recognition of fatty acids by peroxisome proliferator-activated receptors.
Mol.Cell, 3, 1999
|
|
2PAC
 
 | | SOLUTION STRUCTURE OF FE(II) CYTOCHROME C551 FROM PSEUDOMONAS AERUGINOSA AS DETERMINED BY TWO-DIMENSIONAL 1H NMR | | Descriptor: | CYTOCHROME C551, HEME C | | Authors: | Detlefsen, D.J, Thanabal, V, Pecoraro, V.L, Wagner, G. | | Deposit date: | 1993-05-05 | | Release date: | 1993-10-31 | | Last modified: | 2021-03-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Fe(II) cytochrome c551 from Pseudomonas aeruginosa as determined by two-dimensional 1H NMR.
Biochemistry, 30, 1991
|
|
2PIK
 
 | | CALICHEAMICIN GAMMA1I-DNA COMPLEX, NMR, 6 STRUCTURES | | Descriptor: | 2,4-dideoxy-4-(ethylamino)-3-O-methyl-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D-glucopyranose, 2,6-dideoxy-4-thio-beta-D-allopyranose, 3-O-methyl-alpha-L-rhamnopyranose, ... | | Authors: | Kumar, R.A, Ikemoto, N, Patel, D.J. | | Deposit date: | 1996-12-31 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the calicheamicin gamma 1I-DNA complex.
J.Mol.Biol., 265, 1997
|
|
2PTW
 
 | | Crystal Structure of the T. brucei enolase complexed with sulphate, identification of a metal binding site IV | | Descriptor: | 1,2-ETHANEDIOL, Enolase, SULFATE ION, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
2HXM
 
 | | Complex of UNG2 and a small Molecule synthetic Inhibitor | | Descriptor: | 4-[(1E,7E)-8-(2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDIN-4-YL)-3,6-DIOXA-2,7-DIAZAOCTA-1,7-DIEN-1-YL]BENZOIC ACID, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Ghung, S, Seiple, L, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2006-08-03 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mimicking damaged DNA with a small molecule inhibitor of human UNG2.
Nucleic Acids Res., 34, 2006
|
|
4LTS
 
 | | Discovery of Potent and Efficacious Cyanoguanidine-containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 2-cyano-1-pyridin-4-yl-3-(4-{[3-(trifluoromethoxy)phenyl]sulfonyl}benzyl)guanidine, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynoids, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Oh, A, Wang, W, Zak, M, Wang, L, Yuen, P, Bair, K.W. | | Deposit date: | 2013-07-23 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Discovery of potent and efficacious cyanoguanidine-containing nicotinamide phosphoribosyltransferase (Nampt) inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4LWW
 
 | | Discovery of Potent and Efficacious Cyanoguanidine-containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-(4-(phenylsulfonyl)benzyl)-1H-pyrrolo[3,2-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynoids, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Oh, A, Wang, W, Zak, M, Wang, L, Yuen, P, Bair, K.W. | | Deposit date: | 2013-07-28 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Discovery of potent and efficacious cyanoguanidine-containing nicotinamide phosphoribosyltransferase (Nampt) inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3EYJ
 
 | | Structure of Influenza Haemagglutinin in complex with an inhibitor of membrane fusion | | Descriptor: | Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Russell, R.J, Kerry, P.S, Stevens, D.J, Steinahuer, D.A, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2008-10-21 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EYK
 
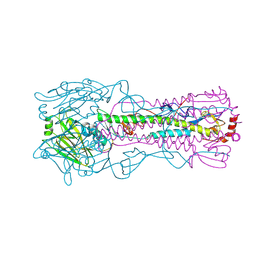 | | Structure of Influenza Haemagglutinin in complex with an inhibitor of membrane fusion | | Descriptor: | 2-tert-butylbenzene-1,4-diol, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Russell, R.J, Kerry, P.S, Stevens, D.J, Steinhauer, D.A, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2008-10-21 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3TNU
 
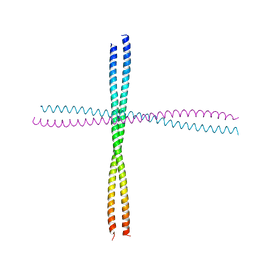 | | Heterocomplex of coil 2B domains of human intermediate filament proteins, keratin 5 (KRT5) and keratin 14 (KRT14) | | Descriptor: | Keratin, type I cytoskeletal 14, type II cytoskeletal 5 | | Authors: | Lee, C.H, Kim, M.S, Leahy, D.J, Coulombe, P.A. | | Deposit date: | 2011-09-02 | | Release date: | 2012-06-20 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structural basis for heteromeric assembly and perinuclear organization of keratin filaments.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3TOR
 
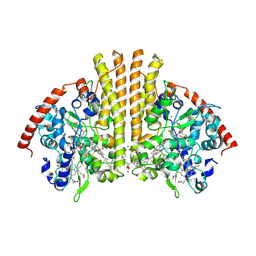 | | Crystal structure of Escherichia coli NrfA with Europium bound | | Descriptor: | CALCIUM ION, Cytochrome c nitrite reductase, EUROPIUM ION, ... | | Authors: | Lockwood, C.W.J, Clarke, T.A, Butt, J.N, Hemmings, A.M, Richardson, D.J. | | Deposit date: | 2011-09-06 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the active site and calcium binding in cytochrome c nitrite reductases.
Biochem.Soc.Trans., 39, 2011
|
|
3U3F
 
 | | Structural basis for the interaction of Pyk2 PAT domain with paxillin LD motifs | | Descriptor: | Paxillin LD2 peptide, Protein-tyrosine kinase 2-beta | | Authors: | Vanarotti, M, Miller, D.J, Guibao, C.C, Zheng, J.J. | | Deposit date: | 2011-10-05 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Structural and Mechanistic Insights into the Interaction between Pyk2 and Paxillin LD Motifs.
J.Mol.Biol., 426, 2014
|
|
3U5P
 
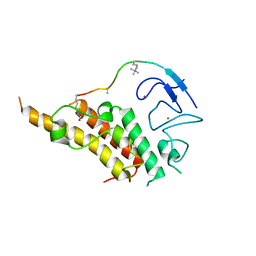 | |
3U9U
 
 | | Crystal Structure of Extracellular Domain of Human ErbB4/Her4 in complex with the Fab fragment of mAb1479 | | Descriptor: | Fab Heavy Chain, Fab Light Chain, Receptor tyrosine-protein kinase erbB-4 | | Authors: | Hollmen, M, Liu, P, Wildiers, H, Reinvall, I, Vandorpe, T, Smeets, A, Deraedt, K, Vahlberg, T, Joensuu, H, Leahy, D.J, Schoffski, P, Elenius, K. | | Deposit date: | 2011-10-19 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Proteolytic processing of ErbB4 in breast cancer.
Plos One, 7, 2012
|
|
3RW7
 
 | | Structure of N-terminal domain of nuclear RNA export factor TAP | | Descriptor: | Nuclear RNA export factor 1 | | Authors: | Teplova, M, Khin, N.W, Wohlbold, L, Izaurralde, E, Patel, D.J. | | Deposit date: | 2011-05-07 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-function studies of nucleocytoplasmic transport of retroviral genomic RNA by mRNA export factor TAP.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3S0I
 
 | | Crystal Structure of D48V mutant of Human Glycolipid Transfer Protein complexed with 3-O-sulfo galactosylceramide containing nervonoyl acyl chain | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Popov, A.N, Cabo-Bilbao, A, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-13 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
