7BBH
 
 | | Structure of Coronavirus Spike from Smuggled Guangdong Pangolin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Surface glycoprotein | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-12-17 | | Release date: | 2020-12-30 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and binding properties of Pangolin-CoV spike glycoprotein inform the evolution of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7A5S
 
 | | Complex of SARS-CoV-2 spike and CR3022 Fab (Homogeneous Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab Heavy Chain, CR3022 Fab Light Chain, ... | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-08-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Antibody-mediated disruption of the SARS-CoV-2 spike glycoprotein.
Nat Commun, 11, 2020
|
|
4P7R
 
 | | Structure of Escherichia coli PgaB C-terminal domain in complex with a poly-beta-1,6-N-acetyl-D-glucosamine (PNAG) hexamer | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5J8H
 
 | | Structure of calmodulin in a complex with a peptide derived from a calmodulin-dependent kinase | | Descriptor: | CALCIUM ION, Calmodulin, Eukaryotic elongation factor 2 kinase | | Authors: | Alphonse, S, Lee, K, Piserchio, A, Tavares, C.D.J, Giles, D.H, Wellmann, R.M, Dalby, K.N, Ghose, R. | | Deposit date: | 2016-04-07 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Recognition of Eukaryotic Elongation Factor 2 Kinase by Calmodulin.
Structure, 24, 2016
|
|
4P7N
 
 | | Structure of Escherichia coli PgaB C-terminal domain in complex with glucosamine | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PEI
 
 | | Dbr1 in complex with synthetic branched RNA analog | | Descriptor: | GLYCEROL, NICKEL (II) ION, RNA (5'-R(*(G46)P*U)-3'), ... | | Authors: | Montemayor, E.J, Katolik, A, Clark, N.E, Taylor, A.B, Schuermann, J.P, Combs, D.J, Johnsson, R, Holloway, S.P, Stevens, S.W, Damha, M.J, Hart, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1.
Nucleic Acids Res., 42, 2014
|
|
4PHK
 
 | | The Structural Basis of Differential Inhibition of Human Calpain by Indole and Phenyl alpha-Mercaptoacrylic Acids. The complex with (Z)-3-(4-chlorophenyl)-2-mercaptoacrylic acid | | Descriptor: | (Z)-3-(4-chlorophenyl)-2-mercaptoacrylic acid, CALCIUM ION, Calpain small subunit 1 | | Authors: | Rizkallah, P.J, Allemann, R.K, Adams, S.E, Miller, D.J, Hallett, M.B, Robinson, E. | | Deposit date: | 2014-05-06 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structural basis of differential inhibition of human calpain by indole and phenyl alpha-mercaptoacrylic acids.
J.Struct.Biol., 187, 2014
|
|
7A92
 
 | | Dissociated S1 domain of SARS-CoV-2 Spike bound to ACE2 (Unmasked Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-30 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A94
 
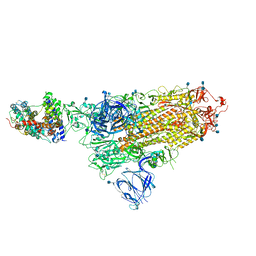 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
4N28
 
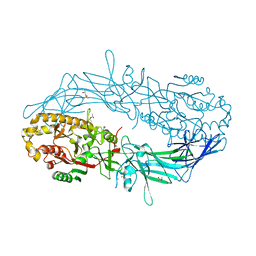 | | Crystal structure of Protein Arginine Deiminase 2 (1 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N2G
 
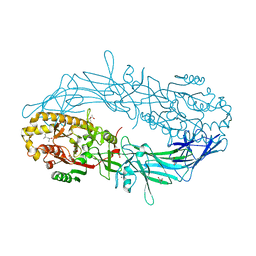 | | Crystal structure of Protein Arginine Deiminase 2 (D169A, 10 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N4D
 
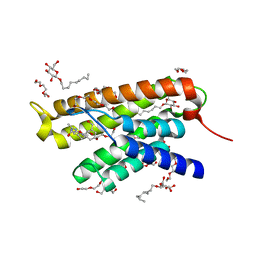 | | Structure of ThiT with AV-38 bound | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Swier, L.J.Y.M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2013-10-08 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies on the thiamine binding protein ThiT
To be Published
|
|
4N2K
 
 | | Crystal structure of Protein Arginine Deiminase 2 (Q350A, 0 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-05 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
7A5R
 
 | | Complex of SARS-CoV-2 spike and CR3022 Fab (Non-Uniform Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab Heavy Chain, CR3022 Fab Light Chain, ... | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-08-21 | | Release date: | 2020-09-16 | | Last modified: | 2020-11-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Antibody-mediated disruption of the SARS-CoV-2 spike glycoprotein.
Nat Commun, 11, 2020
|
|
7A91
 
 | | Dissociated S1 domain of SARS-CoV-2 Spike bound to ACE2 (Non-Uniform Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A95
 
 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound and 1 RBD Erect in Clockwise Direction | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7BNM
 
 | | Closed conformation of D614G SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4NGD
 
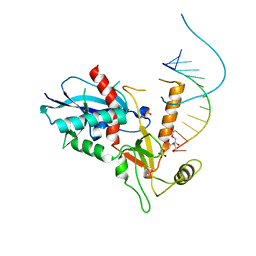 | | Structure of human Dicer Platform-PAZ-Connector Helix cassette in complex with 12-mer siRNA having 5'-p and UU-3' ends (1.95 Angstrom resolution) | | Descriptor: | 5'-R(P*GP*CP*GP*AP*AP*UP*UP*CP*GP*CP*UP*U)-3', DI(HYDROXYETHYL)ETHER, Endoribonuclease Dicer, ... | | Authors: | Tian, Y, Simanshu, D.K, Patel, D.J. | | Deposit date: | 2013-11-01 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | A Phosphate-Binding Pocket within the Platform-PAZ-Connector Helix Cassette of Human Dicer.
Mol.Cell, 53, 2014
|
|
7BNO
 
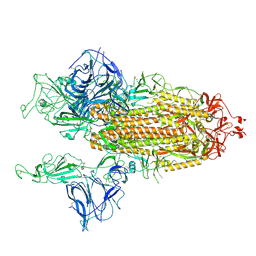 | | Open conformation of D614G SARS-CoV-2 spike with 2 Erect RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BLR
 
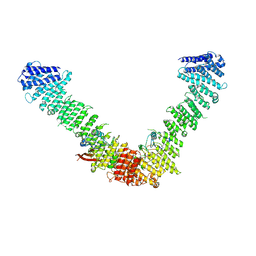 | | Vps35/Vps29 arch of fungal membrane-assembled retromer:Vps5 (SNX-BAR) complex. | | Descriptor: | Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Leneva, N, Kovtun, O, Morado, D.R, Briggs, J.A.G, Owen, D.J. | | Deposit date: | 2021-01-18 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly.
Sci Adv, 7, 2021
|
|
7BLP
 
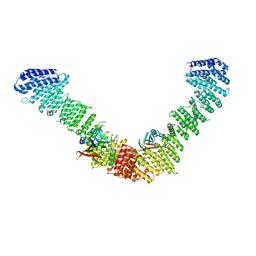 | | Vps35/Vps29 arch of fungal membrane-assembled retromer:Grd19 complex | | Descriptor: | Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Leneva, N, Kovtun, O, Morado, D.R, Briggs, J.A.G, Owen, D.J. | | Deposit date: | 2021-01-18 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly.
Sci Adv, 7, 2021
|
|
7BLN
 
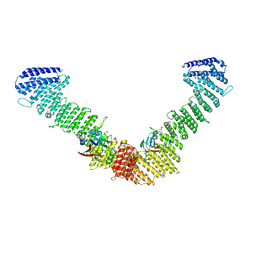 | | VPS35/VPS29 arch of metazoan membrane-assembled retromer:SNX3 complex modelled with human proteins | | Descriptor: | Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Leneva, N, Kovtun, O, Morado, D.R, Briggs, J.A.G, Owen, D.J. | | Deposit date: | 2021-01-18 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly.
Sci Adv, 7, 2021
|
|
4PEG
 
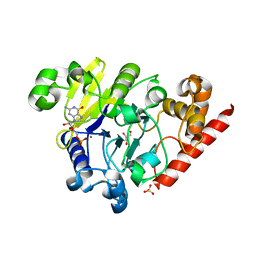 | | Dbr1 in complex with guanosine-5'-monophosphate | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Montemayor, E.J, Katolik, A, Clark, N.E, Taylor, A.B, Schuermann, J.P, Combs, D.J, Johnsson, R, Holloway, S.P, Stevens, S.W, Damha, M.J, Hart, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1.
Nucleic Acids Res., 42, 2014
|
|
7BLO
 
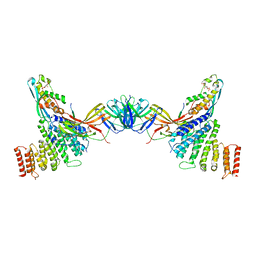 | | VPS26 dimer region of metazoan membrane-assembled retromer:SNX3 complex modelled with human proteins | | Descriptor: | 2-(BUTANOYLOXY)-1-{[(HYDROXY{[2,3,4,6-TETRAHYDROXY-5-(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTANOATE, C-term (residues 493-54) of Wls (fitted sequence corresponds to hDMT1-II), Sorting nexin-3, ... | | Authors: | Leneva, N, Kovtun, O, Morado, D.R, Briggs, J.A.G, Owen, D.J. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly.
Sci Adv, 7, 2021
|
|
4PFJ
 
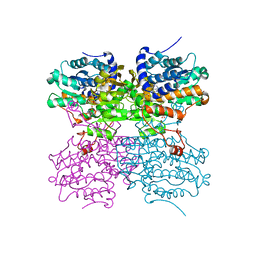 | | The structure of bi-acetylated SAHH | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kavran, J.M, Wang, Y, Cole, P.A, Leahy, D.J. | | Deposit date: | 2014-04-29 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulation of s-adenosylhomocysteine hydrolase by lysine acetylation.
J.Biol.Chem., 289, 2014
|
|
