3FWO
 
 | | The large ribosomal subunit from Deinococcus radiodurans complexed with Methymycin | | Descriptor: | (3R,4S,5S,7R,9E,11S,12R)-12-ethyl-11-hydroxy-3,5,7,11-tetramethyl-2,8-dioxooxacyclododec-9-en-4-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 23S RIBOSOMAL RNA, 5S RIBOSOMAL RNA | | Authors: | Auerbach, T, Mermershtain, I, Bashan, A, Davidovich, C, Rozenberg, H, Sherman, D.H, Yonath, A. | | Deposit date: | 2009-01-19 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Structural basis for the antibacterial activity of the 12-membered-ring mono-sugar macrolide methymycin
Biotechnologia, 1, 2009
|
|
5U2O
 
 | | Crystal structure of Zn-binding triple mutant of GH family 9 endoglucanase J30 | | Descriptor: | CITRATE ANION, GLYCEROL, J30 CCH, ... | | Authors: | Ellinghaus, T.L, Pereira, J.H, McAndrew, R.P, Welner, D.H, Adams, P.D. | | Deposit date: | 2016-11-30 | | Release date: | 2018-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Engineering glycoside hydrolase stability by the introduction of zinc binding.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4NNY
 
 | | Crystal structure of non-phosphorylated form of PKD2 phosphopeptide bound to HLA-A2 | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, GLYCEROL, ... | | Authors: | Mohammed, F, Stones, D.H, Willcox, B.E. | | Deposit date: | 2013-11-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The antigenic identity of human class I MHC phosphopeptides is critically dependent upon phosphorylation status.
Oncotarget, 8, 2017
|
|
2FWO
 
 | |
4NO2
 
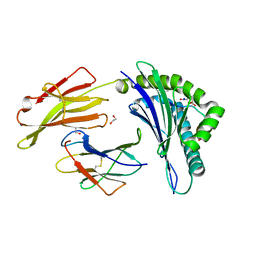 | | Crystal structure of RQA_V phosphopeptide bound to HLA-A2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mohammed, F, Stones, D.H, Willcox, B.E. | | Deposit date: | 2013-11-19 | | Release date: | 2014-11-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The antigenic identity of human class I MHC phosphopeptides is critically dependent upon phosphorylation status.
Oncotarget, 8, 2017
|
|
3I46
 
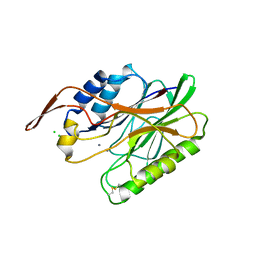 | | Crystal structure of beta toxin from Staphylococcus aureus F277A, P278A mutant with bound calcium ions | | Descriptor: | Beta-hemolysin, CALCIUM ION, CHLORIDE ION | | Authors: | Huseby, M, Shi, K, Kruse, A.C, Ohlendorf, D.H. | | Deposit date: | 2009-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and biological functions of beta toxin from Staphylococcus aureus: Role of the hydrophobic beta hairpin in virulence
To be Published
|
|
4PDC
 
 | |
3I50
 
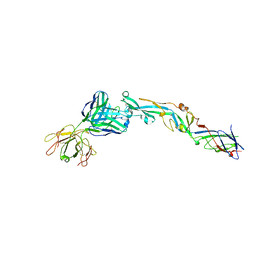 | | Crystal structure of the West Nile Virus envelope glycoprotein in complex with the E53 antibody Fab | | Descriptor: | Envelope glycoprotein, murine heavy chain (IgG3) of E53 monoclonal antibody Fab, murine kappa light chain of E53 monoclonal antibody Fab | | Authors: | Nybakken, G.E, Warren, J.T, Chen, B.R, Nelson, C.A, Fremont, D.H. | | Deposit date: | 2009-07-03 | | Release date: | 2009-10-27 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody.
Embo J., 28, 2009
|
|
3FXI
 
 | | Crystal structure of the human TLR4-human MD-2-E.coli LPS Ra complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-HYDROXY-TETRADECANOIC ACID, ... | | Authors: | Park, B.S, Song, D.H, Kim, H.M, Lee, J.-O. | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex
Nature, 458, 2009
|
|
4MQM
 
 | | Crystal structure of Antigen 85C in presence of Ebselen | | Descriptor: | Diacylglycerol acyltransferase/mycolyltransferase Ag85C | | Authors: | Favrot, L, Grzegorzewicz, A.E, Lajiness, D.H, Marvin, R.K, Boucau, J, Isailovic, D, Jackson, M, Ronning, D.R. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.346 Å) | | Cite: | Mechanism of inhibition of Mycobacterium tuberculosis antigen 85 by ebselen.
Nat Commun, 4, 2013
|
|
5W7P
 
 | | Crystal structure of OxaC | | Descriptor: | OxaC, S-ADENOSYLMETHIONINE | | Authors: | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | Deposit date: | 2017-06-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
3I0Q
 
 | | Crystal Structure of the AMP-bound complex of Spectinomycin Phosphotransferase, APH(9)-Ia | | Descriptor: | ADENOSINE MONOPHOSPHATE, NICKEL (II) ION, Spectinomycin phosphotransferase | | Authors: | Berghuis, A.M, Fong, D.H, Lemke, C.T, Hwang, J.-Y, Xiong, B. | | Deposit date: | 2009-06-25 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila.
J.Biol.Chem., 285, 2010
|
|
3HMP
 
 | | Crystal structure of human Mps1 catalytic domain in complex with a quinazolin ligand Compound 4 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 7-chloro-N-(cyclopropylmethyl)quinazolin-4-amine, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chu, M.L.H, Chavas, L.M.G, Williams, D.H, Tabernero, L, Eyers, P.A. | | Deposit date: | 2009-05-29 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biophysical and X-ray crystallographic analysis of Mps1 kinase inhibitor complexes.
Biochemistry, 49, 2010
|
|
5W7K
 
 | | Crystal structure of OxaG | | Descriptor: | CHLORIDE ION, OxaG, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | Deposit date: | 2017-06-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
5W7S
 
 | | Crystal structure of OxaC in complex with sinefungin and meleagrin | | Descriptor: | (3E,7aR,12aS)-6-hydroxy-3-[(1H-imidazol-4-yl)methylidene]-12-methoxy-7a-(2-methylbut-3-en-2-yl)-7a,12-dihydro-1H,5H-imidazo[1',2':1,2]pyrido[2,3-b]indole-2,5(3H)-dione, OxaC, SINEFUNGIN | | Authors: | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | Deposit date: | 2017-06-20 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.948 Å) | | Cite: | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
4P2P
 
 | | AN INDEPENDENT CRYSTALLOGRAPHIC REFINEMENT OF PORCINE PHOSPHOLIPASE A2 AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Finzel, B.C, Ohlendorf, D.H, Weber, P.C, Salemme, F.R. | | Deposit date: | 1991-10-22 | | Release date: | 1992-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An independent crystallographic refinement of porcine phospholipase A2 at 2.4 A resolution
Acta Crystallogr.,Sect.B, 47, 1991
|
|
5UUA
 
 | |
2GRK
 
 | |
5V2R
 
 | | Structure of a GA Rich 8x8 Nucleotide RNA Internal Loop | | Descriptor: | RNA (5'-R(*CP*CP*AP*GP*AP*AP*AP*CP*GP*GP*AP*UP*GP*GP*A)-3') | | Authors: | Kauffmann, A.D, Kennedy, S.D, Turner, D.H. | | Deposit date: | 2017-03-06 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure of an 8 8 Nucleotide RNA Internal Loop Flanked on Each Side by Three Watson-Crick Pairs and Comparison to Three-Dimensional Predictions.
Biochemistry, 56, 2017
|
|
3HXO
 
 | | Crystal Structure of Von Willebrand Factor (VWF) A1 Domain in Complex with DNA Aptamer ARC1172, an Inhibitor of VWF-Platelet Binding | | Descriptor: | Aptamer ARC1172, von Willebrand factor | | Authors: | Huang, R.H, Sadler, J.E, Fremont, D.H, Diener, J.L, Schaub, R.G. | | Deposit date: | 2009-06-21 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural explanation for the antithrombotic activity of ARC1172, a DNA aptamer that binds von Willebrand factor domain A1.
Structure, 17, 2009
|
|
3I41
 
 | |
3I48
 
 | | Crystal structure of beta toxin from Staphylococcus aureus F277A, P278A mutant with bound magnesium ions | | Descriptor: | Beta-hemolysin, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Huseby, M, Shi, K, Kruse, A.C, Ohlendorf, D.H. | | Deposit date: | 2009-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and biological functions of beta toxin from Staphylococcus aureus: Role of the hydrophobic beta hairpin in virulence
to be published, 2009
|
|
3I5V
 
 | |
4OW1
 
 | | Crystal Structure of Resuscitation Promoting Factor C | | Descriptor: | 1,2-ETHANEDIOL, Resuscitation-promoting factor RpfC | | Authors: | Chauviac, F.X, Quay, D.H.X, Cohen-Gonsaud, M, Keep, N.H. | | Deposit date: | 2014-01-29 | | Release date: | 2014-06-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The RpfC (Rv1884) atomic structure shows high structural conservation within the resuscitation-promoting factor catalytic domain.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
2HG0
 
 | | Structure of the West Nile Virus envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein | | Authors: | Nybakken, G.E, Nelson, C.A, Chen, B.R, Diamond, M.S, Fremont, D.H. | | Deposit date: | 2006-06-26 | | Release date: | 2006-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the West Nile virus envelope glycoprotein.
J.Virol., 80, 2006
|
|
