7Z15
 
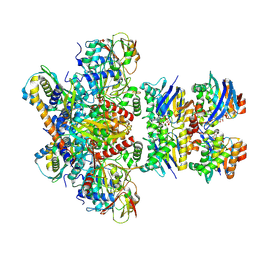 | | E. coli C-P lyase bound to a PhnK/PhnL dual ABC dimer and ADP + Pi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
1P1V
 
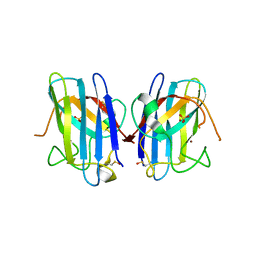 | | Crystal Structure of FALS-associated human Copper-Zinc Superoxide Dismutase (CuZnSOD) Mutant D125H to 1.4A | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Elam, J.S, Malek, K, Rodriguez, J.A, Doucette, P.A, Taylor, A.B, Hayward, L.J, Cabelli, D.E, Valentine, J.S, Hart, P.J. | | Deposit date: | 2003-04-14 | | Release date: | 2003-08-26 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Alternative Mechanism of Bicarbonate-mediated Peroxidation by Copper-Zinc Superoxide Dismutase: RATES ENHANCED VIA PROPOSED ENZYME-ASSOCIATED PEROXYCARBONATE INTERMEDIATE
J.Biol.Chem., 278, 2003
|
|
1P74
 
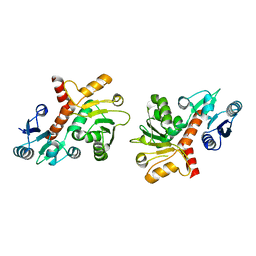 | | CRYSTAL STRUCTURE OF SHIKIMATE DEHYDROGENASE (AROE) FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | Shikimate 5-dehydrogenase | | Authors: | Ye, S, von Delft, F, Brooun, A, Knuth, M.W, Swanson, R.V, McRee, D.E. | | Deposit date: | 2003-04-30 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of shikimate dehydrogenase (AroE) reveals a unique NADPH binding mode
J.Bacteriol., 185, 2003
|
|
1P8O
 
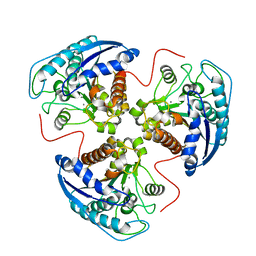 | | Structural and Functional Importance of First-Shell Metal Ligands in the Binuclear Manganese Cluster of Arginase I. | | Descriptor: | Arginase 1, MANGANESE (II) ION | | Authors: | Cama, E, Emig, F.A, Ash, D.E, Christianson, D.W. | | Deposit date: | 2003-05-07 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structural and functional importance of first-shell metal ligands in the binuclear
manganese cluster of arginase I
Biochemistry, 42, 2003
|
|
1HQ5
 
 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH S-(2-BORONOETHYL)-L-CYSTEINE, AN L-ARGININE ANALOGUE | | Descriptor: | ARGINASE 1, MANGANESE (II) ION, S-2-(BORONOETHYL)-L-CYSTEINE | | Authors: | Kim, N.N, Cox, J.D, Baggio, R.F, Emig, F.A, Mistry, S.K, Harper, S.L, Speicher, D.W, Morris Jr, S.M, Ash, D.E, Traish, A, Christianson, D.W. | | Deposit date: | 2000-12-14 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing erectile function: S-(2-boronoethyl)-L-cysteine binds to arginase as a transition state analogue and enhances smooth muscle relaxation in human penile corpus cavernosum.
Biochemistry, 40, 2001
|
|
1HQH
 
 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH NOR-N-HYDROXY-L-ARGININE | | Descriptor: | ARGINASE 1, MANGANESE (II) ION, NOR-N-OMEGA-HYDROXY-L-ARGININE | | Authors: | Cox, J.D, Cama, E, Colleluori, D.M, Ash, D.E, Christianson, D.W. | | Deposit date: | 2000-12-16 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and metabolic inferences from the binding of substrate analogues and products to arginase.
Biochemistry, 40, 2001
|
|
1OZO
 
 | | Three-dimensional solution structure of apo-S100P protein determined by NMR spectroscopy | | Descriptor: | S-100P protein | | Authors: | Lee, Y.-C, Volk, D.E, Thiviyanathan, V, Kleerekoper, Q, Gribenko, A.V, Zhang, S, Gorenstein, D.G, Makhatadze, G.I, Luxon, B.A. | | Deposit date: | 2003-04-09 | | Release date: | 2004-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Apo-S100P protein.
J.Biomol.Nmr, 29, 2004
|
|
4TMN
 
 | | SLOW-AND FAST-BINDING INHIBITORS OF THERMOLYSIN DISPLAY DIFFERENT MODES OF BINDING. CRYSTALLOGRAPHIC ANALYSIS OF EXTENDED PHOSPHONAMIDATE TRANSITION-STATE ANALOGUES | | Descriptor: | CALCIUM ION, N-[(S)-[(1R)-1-{[(benzyloxy)carbonyl]amino}-2-phenylethyl](hydroxy)phosphoryl]-L-leucyl-L-alanine, THERMOLYSIN, ... | | Authors: | Holden, H.M, Tronrud, D.E, Monzingo, A.F, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Slow- and fast-binding inhibitors of thermolysin display different modes of binding: crystallographic analysis of extended phosphonamidate transition-state analogues.
Biochemistry, 26, 1987
|
|
4TNH
 
 | | RT XFEL structure of Photosystem II in the dark state at 4.9 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.900007 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
4TNL
 
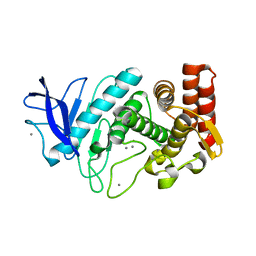 | | 1.8 A resolution room temperature structure of Thermolysin recorded using an XFEL | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
1R6X
 
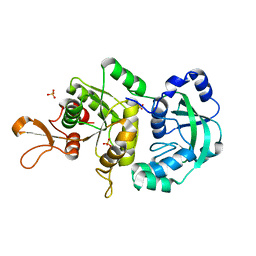 | | The Crystal Structure of a Truncated Form of Yeast ATP Sulfurylase, Lacking the C-Terminal APS Kinase-like Domain, in complex with Sulfate | | Descriptor: | ATP:sulfate adenylyltransferase, COBALT (II) ION, SULFATE ION | | Authors: | Lalor, D.J, Schnyder, T, Saridakis, V, Pilloff, D.E, Dong, A, Tang, H, Leyh, T.S, Pai, E.F. | | Deposit date: | 2003-10-17 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional analysis of a truncated form of Saccharomyces cerevisiae ATP sulfurylase: C-terminal domain essential for oligomer formation but not for activity
Protein Eng., 16, 2003
|
|
1R4I
 
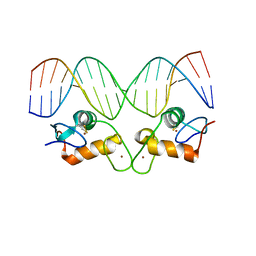 | | Crystal Structure of Androgen Receptor DNA-Binding Domain Bound to a Direct Repeat Response Element | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*TP*CP*AP*AP*GP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*TP*CP*TP*TP*GP*AP*TP*GP*TP*TP*CP*TP*GP*G)-3', Androgen receptor, ... | | Authors: | Shaffer, P.L, Jivan, A, Dollins, D.E, Claessens, F, Gewirth, D.T. | | Deposit date: | 2003-10-06 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of androgen receptor binding to selective androgen response elements.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1OJD
 
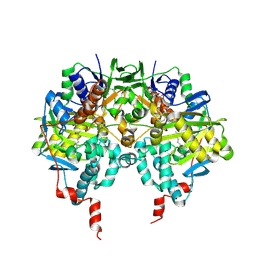 | |
1OJA
 
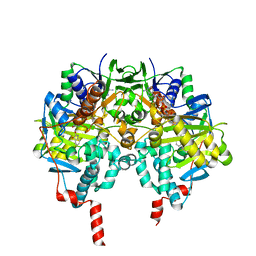 | | HUMAN MONOAMINE OXIDASE B IN COMPLEX WITH ISATIN | | Descriptor: | AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE, ISATIN | | Authors: | Binda, C, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2003-07-08 | | Release date: | 2003-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights Into the Mode of Inhibition of Human Mitochondrial Monoamine Oxidase B from High-Resolution Crystal Structures
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1OJ9
 
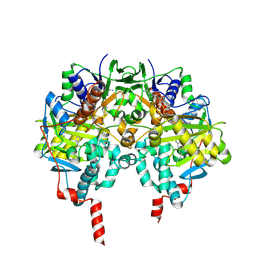 | | HUMAN MONOAMINE OXIDASE B IN COMPLEX WITH 1,4-DIPHENYL-2-BUTENE | | Descriptor: | 1,4-DIPHENYL-2-BUTENE, AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2003-07-08 | | Release date: | 2003-08-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights Into the Mode of Inhibition of Human Mitochondrial Monoamine Oxidase B from High-Resolution Crystal Structures
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1MT1
 
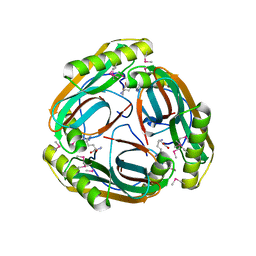 | | The Crystal Structure of Pyruvoyl-dependent Arginine Decarboxylase from Methanococcus jannaschii | | Descriptor: | AGMATINE, PYRUVOYL-DEPENDENT ARGININE DECARBOXYLASE ALPHA CHAIN, PYRUVOYL-DEPENDENT ARGININE DECARBOXYLASE BETA CHAIN | | Authors: | Tolbert, W.D, Graham, D.E, White, R.H, Ealick, S.E. | | Deposit date: | 2002-09-20 | | Release date: | 2003-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii:
Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms
Structure, 11, 2003
|
|
1R6T
 
 | | crystal structure of human tryptophanyl-tRNA synthetase | | Descriptor: | GLYCEROL, TRYPTOPHANYL-5'AMP, Tryptophanyl-tRNA synthetase | | Authors: | Yang, X.-L, Otero, F.J, Skene, R.J, McRee, D.E, Ribas de Pouplana, L, Schimmel, P. | | Deposit date: | 2003-10-16 | | Release date: | 2004-01-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1R6U
 
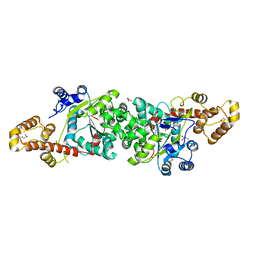 | | Crystal structure of an active fragment of human tryptophanyl-tRNA synthetase with cytokine activity | | Descriptor: | GLYCEROL, TRYPTOPHANYL-5'AMP, Tryptophanyl-tRNA synthetase | | Authors: | Yang, X.-L, Otero, F.J, Skene, R.J, McRee, D.E, Ribas de Pouplana, L, Schimmel, P. | | Deposit date: | 2003-10-16 | | Release date: | 2004-01-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and crystal structure analysis of active site adaptations of a potent anti-angiogenic human tRNA synthetase
Structure, 15, 2007
|
|
1FVT
 
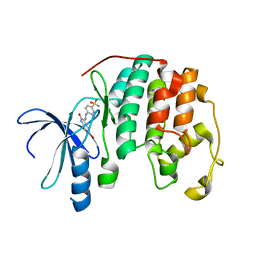 | | THE STRUCTURE OF CYCLIN-DEPENDENT KINASE 2 (CDK2) IN COMPLEX WITH AN OXINDOLE INHIBITOR | | Descriptor: | 4-[(2Z)-2-(5-bromo-2-oxo-1,2-dihydro-3H-indol-3-ylidene)hydrazinyl]benzene-1-sulfonamide, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Davis, S.T, Benson, B.G, Bramson, H.N, Chapman, D.E, Dickerson, S.H, Dold, K.M, Eberwein, D.J, Edelstein, M, Frye, S.V, Gampe Jr, R.T, Griffin, R.J, Harris, P.A, Hassell, A.M, Holmes, W.D, Hunter, R.N, Knick, V.B, Lackey, K, Lovejoy, B, Luzzio, M.J, Murray, D, Parker, P, Rocque, W.J, Shewchuk, L, Veal, J.M, Walker, D.H, Kuyper, L.K. | | Deposit date: | 2000-09-20 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prevention of chemotherapy-induced alopecia in rats by CDK inhibitors.
Science, 291, 2001
|
|
1N13
 
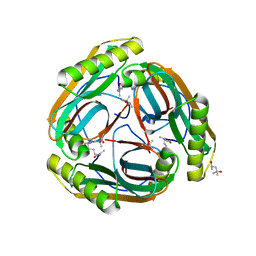 | | The Crystal Structure of Pyruvoyl-dependent Arginine Decarboxylase from Methanococcus jannashii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, AGMATINE, Pyruvoyl-dependent arginine decarboxylase alpha chain, ... | | Authors: | Tolbert, W.D, Graham, D.E, White, R.H, Ealick, S.E. | | Deposit date: | 2002-10-16 | | Release date: | 2003-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii:
Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms
Structure, 11, 2003
|
|
1Q35
 
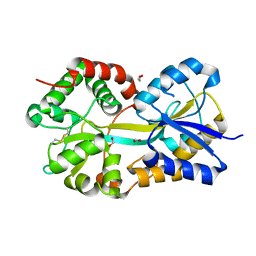 | | Crystal Structure of Pasteurella haemolytica Apo Ferric ion-Binding Protein A | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, iron binding protein FbpA | | Authors: | Shouldice, S.R, Dougan, D.R, Skene, R.J, Snell, G, Scheibe, D, Williams, P.A, Kirby, S, McRee, D.E, Schryvers, A.B, Tari, L.W. | | Deposit date: | 2003-07-28 | | Release date: | 2003-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of Pasteurella haemolytica ferric ion-binding protein A reveals a novel class of bacterial iron-binding proteins
J.Biol.Chem., 278, 2003
|
|
1FYY
 
 | | HPRT GENE MUTATION HOTSPOT WITH A BPDE2(10R) ADDUCT | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 5'-D(*TP*GP*CP*CP*CP*TP*TP*GP*AP*CP*TP*A)-3', HPRT DNA WITH BENZO[A]PYRENE-ADDUCTED DA7 | | Authors: | Volk, D.E, Rice, J.S, Luxon, B.A, Yeh, H.J.C, Liang, C, Xie, G, Sayer, J.M, Jerina, D.M, Gorenstein, D.G. | | Deposit date: | 2000-10-03 | | Release date: | 2000-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR evidence for syn-anti interconversion of a trans opened (10R)-dA adduct of benzo[a]pyrene (7S,8R)-diol (9R,10S)-epoxide in a DNA duplex.
Biochemistry, 39, 2000
|
|
1PTM
 
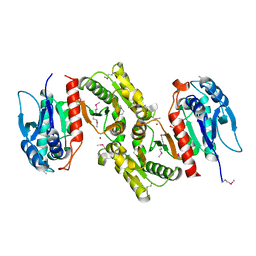 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, PHOSPHATE ION, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-06-23 | | Release date: | 2003-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
1N2M
 
 | | The S53A Proenzyme Structure of Methanococcus jannaschii. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Pyruvoyl-dependent arginine decarboxylase | | Authors: | Tolbert, W.D, Graham, D.E, White, R.H, Ealick, S.E. | | Deposit date: | 2002-10-23 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii:
Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms
Structure, 11, 2003
|
|
1RLM
 
 | | Crystal Structure of ybiV from Escherichia coli K12 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphatase | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
