6WA2
 
 | | Crystal structure of EGFR(T790M/V948R) in complex with LN3753 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N-(3-{5-[2-(acetylamino)pyridin-4-yl]-2-(methylsulfanyl)-1H-imidazol-4-yl}phenyl)-2-fluoro-5-hydroxybenzamide | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of a "Two-in-One" Mutant-Selective Epidermal Growth Factor Receptor Inhibitor That Spans the Orthosteric and Allosteric Sites.
J.Med.Chem., 65, 2022
|
|
6WAK
 
 | | A crystal structure of EGFR(T790M/V948R) in complex with LN3754 | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, N-(3-{5-[2-(acetylamino)pyridin-4-yl]-2-(methylsulfanyl)-1H-imidazol-4-yl}phenyl)-2-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]benzamide, ... | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of a "Two-in-One" Mutant-Selective Epidermal Growth Factor Receptor Inhibitor That Spans the Orthosteric and Allosteric Sites.
J.Med.Chem., 65, 2022
|
|
5L1Q
 
 | | X-ray Structure of Cytochrome P450 PntM with Dihydropentalenolactone F | | Descriptor: | Dihydropentalenolactone F, PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5L1S
 
 | |
5L1U
 
 | |
5L8Z
 
 | | Structure of thermostable DNA-binding HU protein from micoplasma Spiroplasma melliferum | | Descriptor: | DNA-binding protein, SODIUM ION | | Authors: | Boyko, K.M, Gorbacheva, M.A, Rakitina, T.V, Korzhenevskiy, D.A, Kamashev, D.E, Vanyushkina, A.A, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the high thermal stability of the histone-like HU protein from the mollicute Spiroplasma melliferum KC3.
Sci Rep, 6, 2016
|
|
5L6L
 
 | | Structure of Caulobacter crescentus VapBC1 bound to operator DNA | | Descriptor: | DNA (27-MER), Ribonuclease VapC, VapB family protein | | Authors: | Bendtsen, K.L, Xu, K, Luckmann, M, Brodersen, D.E. | | Deposit date: | 2016-05-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Toxin inhibition in C. crescentus VapBC1 is mediated by a flexible pseudo-palindromic protein motif and modulated by DNA binding.
Nucleic Acids Res., 45, 2017
|
|
5L1T
 
 | |
5L6M
 
 | | Structure of Caulobacter crescentus VapBC1 (VapB1deltaC:VapC1 form) | | Descriptor: | GLYCEROL, MALONATE ION, Ribonuclease VapC, ... | | Authors: | Bendtsen, K.L, Xu, K, Luckmann, M, Brodersen, D.E. | | Deposit date: | 2016-05-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Toxin inhibition in C. crescentus VapBC1 is mediated by a flexible pseudo-palindromic protein motif and modulated by DNA binding.
Nucleic Acids Res., 45, 2017
|
|
3NP1
 
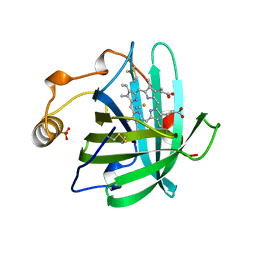 | | CRYSTAL STRUCTURE OF THE COMPLEX OF NITROPHORIN 1 FROM RHODNIUS PROLIXUS WITH CYANIDE | | Descriptor: | CYANIDE ION, NITROPHORIN 1, PHOSPHATE ION, ... | | Authors: | Weichsel, A, Andersen, J.F, Champagne, D.E, Walker, F.A, Montfort, W.R. | | Deposit date: | 1998-01-22 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of a nitric oxide transport protein from a blood-sucking insect.
Nat.Struct.Biol., 5, 1998
|
|
4HNW
 
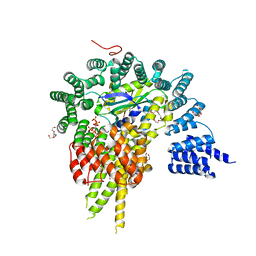 | | The NatA Acetyltransferase Complex Bound To Inositol Hexakisphosphate | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, N-terminal acetyltransferase A complex catalytic subunit ARD1, N-terminal acetyltransferase A complex subunit NAT1, ... | | Authors: | Neubauer, J.L, Immormino, R.M, Dollins, D.E, Endo-Streeter, S.T, Pemble IV, C.W, York, J.D. | | Deposit date: | 2012-10-21 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | The Protein Complex NatA Binds Inositol Hexakisphosphate and Exhibits Conformational Flexibility
To be Published
|
|
1GK5
 
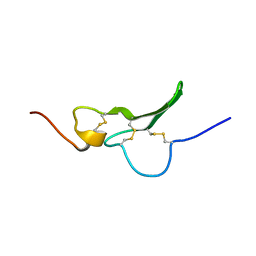 | | Solution Structure the mEGF/TGFalpha44-50 chimeric growth factor | | Descriptor: | Pro-epidermal growth factor,Protransforming growth factor alpha | | Authors: | Chamberlin, S.G, Brennan, L, Puddicombe, S.M, Davies, D.E, Turner, D.L. | | Deposit date: | 2001-08-08 | | Release date: | 2002-08-08 | | Last modified: | 2018-03-28 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Megf/Tgfalpha44-50 Chimeric Growth Factor.
Eur.J.Biochem., 268, 2001
|
|
1LN2
 
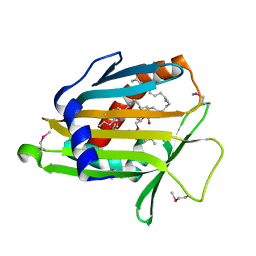 | | Crystal Structure of Human Phosphatidylcholine Transfer Protein in Complex with Dilinoleoylphosphatidylcholine (Seleno-Met Protein) | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Phosphatidylcholine transfer protein | | Authors: | Roderick, S.L, Chan, W.W, Agate, D.S, Olsen, L.R, Vetting, M.W, Rajashankar, K.R, Cohen, D.E. | | Deposit date: | 2002-05-02 | | Release date: | 2002-06-26 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of human phosphatidylcholine transfer protein in complex with its ligand.
Nat.Struct.Biol., 9, 2002
|
|
4HNY
 
 | | Apo N-terminal acetyltransferase complex A | | Descriptor: | GLYCEROL, N-terminal acetyltransferase A complex catalytic subunit ARD1, N-terminal acetyltransferase A complex subunit NAT1, ... | | Authors: | Neubauer, J.L, Immormino, R.M, Dollins, D.E, Endo-Streeter, S.T, Pemble IV, C.W, York, J.D. | | Deposit date: | 2012-10-21 | | Release date: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | The Protein Complex NatA Binds Inositol Hexakisphosphate and Exhibits Conformational Flexibility
To be Published
|
|
4HJD
 
 | | GCN4pLI derivative with alpha/beta/acyclic-gamma amino acid substitution pattern | | Descriptor: | GCN4pLI(alpha/beta/acyclic gamma) | | Authors: | Shin, Y.H, Mortenson, D.E, Satyshur, K.A, Forest, K.T, Gellman, S.H. | | Deposit date: | 2012-10-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Differential Impact of beta and gamma Residue Preorganization on alpha / beta / gamma-Peptide Helix Stability in Water.
J.Am.Chem.Soc., 135, 2013
|
|
3FDQ
 
 | |
4MCN
 
 | | Human SOD1 C57S Mutant, Metal-free | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn] | | Authors: | Sea, K, Sohn, S.H, Durazo, A, Sheng, Y, Shaw, B, Cao, X, Taylor, A.B, Whitson, L.J, Holloway, S.P, Hart, P.J, Cabelli, D.E, Gralla, E.B, Valentine, J.S. | | Deposit date: | 2013-08-21 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the role of the unusual disulfide bond in copper-zinc superoxide dismutase.
J.Biol.Chem., 290, 2015
|
|
4MIT
 
 | |
1LIS
 
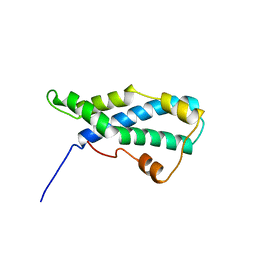 | | THE CRYSTAL STRUCTURE OF A FERTILIZATION PROTEIN | | Descriptor: | LYSIN | | Authors: | Shaw, A, Mcree, D.E, Vacquier, V.D, Stout, C.D. | | Deposit date: | 1993-06-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of lysin, a fertilization protein.
Science, 262, 1993
|
|
4HJB
 
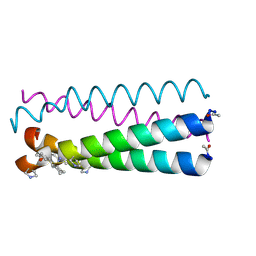 | | GCN4pLI derivative with alpha/beta/cyclic-gamma amino acid substitution pattern | | Descriptor: | GCN4pLI(alpha/beta/cyclic-gamma) | | Authors: | Shin, Y.H, Mortenson, D.E, Satyshur, K.A, Forest, K.T, Gellman, S.H. | | Deposit date: | 2012-10-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Differential Impact of beta and gamma Residue Preorganization on alpha / beta / gamma-Peptide Helix Stability in Water.
J.Am.Chem.Soc., 135, 2013
|
|
1LW6
 
 | |
1MP8
 
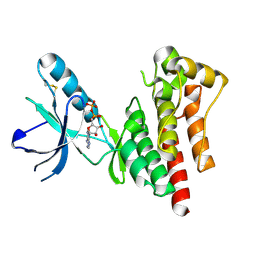 | | Crystal structure of Focal Adhesion Kinase (FAK) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, focal adhesion kinase 1 | | Authors: | Nowakowski, J, Cronin, C.N, McRee, D.E, Knuth, M.W, Nelson, C.G, Pavletich, N.P, Rodgers, J, Sang, B.-C, Scheibe, D.N, Swanson, R.V, Thompson, D.A. | | Deposit date: | 2002-09-11 | | Release date: | 2003-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of the cancer-related Aurora-A, FAK, and EphA2 protein kinases from nanovolume crystallography
Structure, 10, 2002
|
|
1MNA
 
 | | Thioesterase Domain of Picromycin Polyketide Synthase (PICS TE), pH 8.0 | | Descriptor: | polyketide synthase IV | | Authors: | Tsai, S.-C, Lu, H, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2002-09-05 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases
Biochemistry, 41, 2002
|
|
1MNQ
 
 | | Thioesterase Domain of Picromycin Polyketide Synthase (PICS TE), pH 8.4 | | Descriptor: | polyketide synthase IV | | Authors: | Tsai, S.-C, Lu, H, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2002-09-05 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases
Biochemistry, 41, 2002
|
|
3GT8
 
 | | Crystal structure of the inactive EGFR kinase domain in complex with AMP-PNP | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Jura, N, Endres, N.F, Engel, K, Deindl, S, Das, R, Lamers, M.H, Wemmer, D.E, Zhang, X, Kuriyan, J. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment.
Cell(Cambridge,Mass.), 137, 2009
|
|
