4D2T
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 3-[2-(phenylcarbamoyl)-5-(1H-pyrazol-4-yl)phenoxy]propan-1-aminium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-13 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4D2P
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 7-({4-[(3-hydroxy-5-methoxyphenyl)amino]benzoyl}amino)-1,2,3,4-tetrahydroisoquinolinium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-12 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
6BIQ
 
 | |
4CMW
 
 | | Crystal structure of Rv3378c | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Layre, E, Lee, H.J, Young, D.C, Martinot, A.J, Buter, J, Minnaard, A.J, Annand, J.W, Fortune, S.M, Snider, B.B, Matsunaga, I, Rubin, E.J, Alber, T, Moody, D.B. | | Deposit date: | 2014-01-18 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Molecular Profiling of Mycobacterium Tuberculosis Identifies Tuberculosinyl Nucleoside Products of the Virulence-Associated Enzyme Rv3378C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6BIO
 
 | |
4CMX
 
 | | Crystal structure of Rv3378c | | Descriptor: | 1,2-ETHANEDIOL, BENZENE HEXACARBOXYLIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Layre, E, Lee, H.J, Young, D.C, Martinot, A.J, Buter, J, Minnaard, A.J, Annand, J.W, Fortune, S.M, Snider, B.B, Matsunaga, I, Rubin, E.J, Alber, T, Moody, D.B. | | Deposit date: | 2014-01-18 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Molecular Profiling of Mycobacterium Tuberculosis Identifies Tuberculosinyl Nucleoside Products of the Virulence-Associated Enzyme Rv3378C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6BO1
 
 | | Mono-adduct formed after 3 days in the reaction of dichlorido(1,3-dimethylbenzimidazol-2-ylidene)(eta6-p-cymene)ruthenium(II) with HEWL | | Descriptor: | Lysozyme C, SODIUM ION, dichloro(1,3-dimethyl-1H-benzimidazol-3-ium-2-yl)ruthenium | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2017-11-17 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Unexpected arene ligand exchange results in the oxidation of an organoruthenium anticancer agent: the first X-ray structure of a protein-Ru(carbene) adduct.
Chem. Commun. (Camb.), 54, 2018
|
|
6BG3
 
 | | Structure of (3S,4S)-1-benzyl-4-(3-(3-(trifluoromethyl)phenyl)ureido)piperidin-3-yl acetate bound to DCN1 | | Descriptor: | Endolysin, DCN1-like protein 1 chimera, N-{(3S,4S)-1-benzyl-3-[(1S)-1-hydroxyethoxy]piperidin-4-yl}-N'-[3-(trifluoromethyl)phenyl]urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-10-27 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Piperidinyl Ureas Chemically Control Defective in Cullin Neddylation 1 (DCN1)-Mediated Cullin Neddylation.
J. Med. Chem., 61, 2018
|
|
6BIM
 
 | |
6C42
 
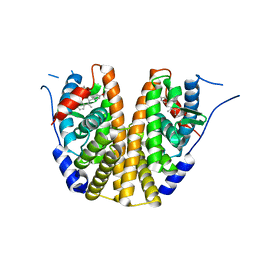 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with OP1156 | | Descriptor: | (2R,3S,4R)-3-(4-hydroxyphenyl)-4-methyl-2-{4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}-3,4-dihydro-2H-1-benzopyran-7-ol, Estrogen receptor | | Authors: | Fanning, S.W, Hodges-Gallager, L, Myles, D.C, Sun, R, Fowler, C.E, Green, B.D, Harmon, C.L, Greene, G.L, Kushner, P.J. | | Deposit date: | 2018-01-11 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Specific stereochemistry of OP-1074 disrupts estrogen receptor alpha helix 12 and confers pure antiestrogenic activity.
Nat Commun, 9, 2018
|
|
6CYM
 
 | |
6CN7
 
 | |
1A6A
 
 | | THE STRUCTURE OF AN INTERMEDIATE IN CLASS II MHC MATURATION: CLIP BOUND TO HLA-DR3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Ghosh, P, Amaya, M, Mellins, E, Wiley, D.C. | | Deposit date: | 1998-02-22 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure of an intermediate in class II MHC maturation: CLIP bound to HLA-DR3.
Nature, 378, 1995
|
|
1AIK
 
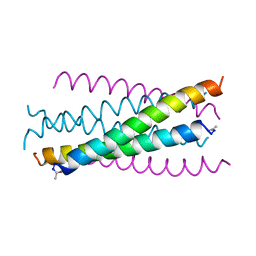 | | HIV GP41 CORE STRUCTURE | | Descriptor: | HIV-1 GP41 GLYCOPROTEIN | | Authors: | Chan, D.C, Fass, D, Berger, J.M, Kim, P.S. | | Deposit date: | 1997-04-20 | | Release date: | 1997-06-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Core structure of gp41 from the HIV envelope glycoprotein.
Cell(Cambridge,Mass.), 89, 1997
|
|
1AMF
 
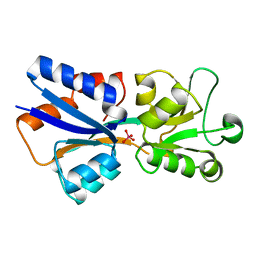 | | CRYSTAL STRUCTURE OF MODA, A MOLYBDATE TRANSPORT PROTEIN, COMPLEXED WITH MOLYBDATE | | Descriptor: | MOLYBDATE ION, MOLYBDATE TRANSPORT PROTEIN MODA | | Authors: | Hu, Y, Rech, S, Gunsalus, R.P, Rees, D.C. | | Deposit date: | 1997-06-13 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the molybdate binding protein ModA.
Nat.Struct.Biol., 4, 1997
|
|
1AG0
 
 | |
1B25
 
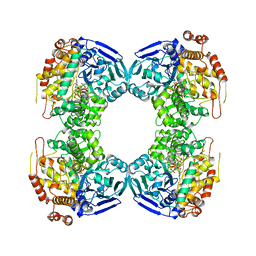 | | FORMALDEHYDE FERREDOXIN OXIDOREDUCTASE FROM PYROCOCCUS FURIOSUS | | Descriptor: | IRON/SULFUR CLUSTER, PROTEIN (FORMALDEHYDE FERREDOXIN OXIDOREDUCTASE), TUNGSTOPTERIN | | Authors: | Hu, Y.L, Faham, S, Roy, R, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1998-12-04 | | Release date: | 1999-03-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Formaldehyde ferredoxin oxidoreductase from Pyrococcus furiosus: the 1.85 A resolution crystal structure and its mechanistic implications.
J.Mol.Biol., 286, 1999
|
|
6DEW
 
 | | Structure of human COQ9 protein with bound isoprene. | | Descriptor: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, (2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, ... | | Authors: | Bingman, C.A, Lohman, D.C, Smith, R.W, Pagliarini, D.J. | | Deposit date: | 2018-05-13 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Isoprene Lipid-Binding Protein Promotes Eukaryotic Coenzyme Q Biosynthesis.
Mol.Cell, 73, 2019
|
|
1B4N
 
 | | FORMALDEHYDE FERREDOXIN OXIDOREDUCTASE FROM PYROCOCCUS FURIOSUS, COMPLEXED WITH GLUTARATE | | Descriptor: | CALCIUM ION, FORMALDEHYDE FERREDOXIN OXIDOREDUCTASE, GLUTARIC ACID, ... | | Authors: | Hu, Y.L, Faham, S, Roy, R, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1998-12-24 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Formaldehyde ferredoxin oxidoreductase from Pyrococcus furiosus: the 1.85 A resolution crystal structure and its mechanistic implications.
J.Mol.Biol., 286, 1999
|
|
8R3T
 
 | | Cofactor-free Tau 4R2N isoform | | Descriptor: | Microtubule-associated protein tau | | Authors: | Limorenko, G, Tatli, M, Kolla, R, Nazarov, S, Weil, M.T, Schondorf, D.C, Geist, D, Reinhardt, P, Ehrnhoefer, D.E, Stahlberg, H, Gasparini, L, Lashuel, H.A. | | Deposit date: | 2023-11-10 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Fully co-factor-free ClearTau platform produces seeding-competent Tau fibrils for reconstructing pathological Tau aggregates.
Nat Commun, 14, 2023
|
|
6DTC
 
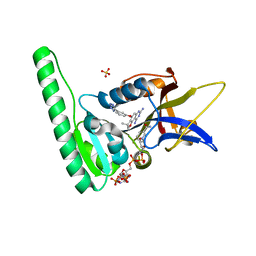 | | Dihydrofolate Reductase (DHFR) of Aspergillus flavus in complex with a small molecule inhibitor | | Descriptor: | (3R)-3-methyl-4-[3-(1H-tetrazol-5-yl)phenoxy]-2,3-dihydrofuro[2,3-f]quinazoline-7,9-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Bensen, D.C, Fortier, J.M, Akers-Rodriguez, S, Tari, L.W. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prospecting for broad-spectrum inhibitors of fungal dihydrofolate reductase using a structure guided approach.
To be published
|
|
1B0R
 
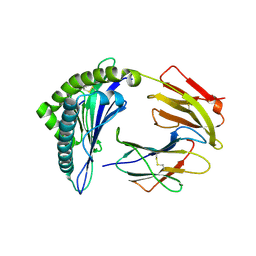 | | CRYSTAL STRUCTURE OF HLA-A*0201 COMPLEXED WITH A PEPTIDE WITH THE CARBOXYL-TERMINAL GROUP SUBSTITUTED BY A METHYL GROUP | | Descriptor: | PROTEIN (HLA-A*0201), PROTEIN (INFLUENZA MATRIX PEPTIDE) | | Authors: | Bouvier, M, Guo, H, Smith, K.J, Wiley, D.C. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of HLA-A*0201 complexed with antigenic peptides with either the amino- or carboxyl-terminal group substituted by a methyl group.
Proteins, 33, 1998
|
|
6DRS
 
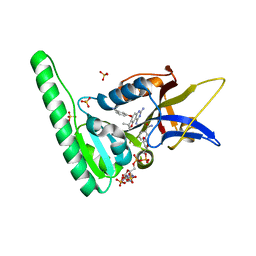 | | Dihydrofolate Reductase (DHFR) of Aspergillus flavus in complex with a small molecule inhibitor | | Descriptor: | 3-{[(3R)-7,9-diamino-3-methyl-2,3-dihydrofuro[2,3-f]quinazolin-4-yl]oxy}benzonitrile, Dihydrofolate reductase, putative, ... | | Authors: | Bensen, D.C, Fortier, J.M, Akers-Rodriguez, S, Tari, L.W. | | Deposit date: | 2018-06-12 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Prospecting for broad-spectrum inhibitors of fungal dihydrofolate reductase using a structure guided approach.
To Be Published
|
|
1B6B
 
 | |
1B9E
 
 | | HUMAN INSULIN MUTANT SERB9GLU | | Descriptor: | PROTEIN (INSULIN) | | Authors: | Wang, D.C, Zeng, Z.H, Yao, Z.P, Li, H.M. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an insulin dimer in an orthorhombic crystal: the structure analysis of a human insulin mutant (B9 Ser-->Glu).
Acta Crystallogr.,Sect.D, 55, 1999
|
|
