6OJA
 
 | | Crystal structure of the N. meningitides methionine-binding protein in its L-methionine bound conformation | | Descriptor: | Lipoprotein, METHIONINE | | Authors: | Nguyen, P.T, Lai, J.Y, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2019-04-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of the Neisseria meningitides methionine-binding protein MetQ in substrate-free form and bound to l- and d-methionine isomers.
Protein Sci., 28, 2019
|
|
6OP3
 
 | |
6OP2
 
 | |
6OP4
 
 | | Selenium-incorporated, carbon monoxide-inhibited, reactivated FeMo-cofactor of nitrogenase from Azotobacter vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Arias, R.J, Rees, D.C. | | Deposit date: | 2019-04-24 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Localized Electronic Structure of Nitrogenase FeMoco Revealed by Selenium K-Edge High Resolution X-ray Absorption Spectroscopy.
J.Am.Chem.Soc., 141, 2019
|
|
2J17
 
 | | pTyr bound form of SDP-1 | | Descriptor: | MAGNESIUM ION, O-PHOSPHOTYROSINE, TYROSINE-PROTEIN PHOSPHATASE YIL113W | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2006-08-09 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Redox-mediated substrate recognition by Sdp1 defines a new group of tyrosine phosphatases.
Nature, 447, 2007
|
|
6QOZ
 
 | | CryoEM reconstruction of Cowpea Mosaic Virus (CPMV) bound to an Affimer reagent | | Descriptor: | Affimer binding protein, Cowpea mosaic virus large subunit, RNA2 polyprotein | | Authors: | Hesketh, E.L, Tiede, C, Adamson, H, Adams, T.L, Byrne, M.J, Meshcheriakova, Y, Lomonossoff, G.P, Kruse, I, McPherson, M.J, Tomlinson, D.C, Ranson, N.A. | | Deposit date: | 2019-02-13 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Affimer reagents as tools in diagnosing plant virus diseases.
Sci Rep, 9, 2019
|
|
6QZK
 
 | | Structure of Clostridium butyricum Argonaute bound to a guide DNA (5' deoxycytidine) and a 19-mer target DNA | | Descriptor: | Clostridium butyricum Argonaute, DNA target (5'-D(T*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*T)-3'), FORMIC ACID, ... | | Authors: | Swarts, D.C, Jinek, M, Hegge, J.W, Van der Oost, J. | | Deposit date: | 2019-03-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.548 Å) | | Cite: | DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute.
Nucleic Acids Res., 47, 2019
|
|
2IYB
 
 | |
2J16
 
 | | Apo & Sulphate bound forms of SDP-1 | | Descriptor: | MAGNESIUM ION, SULFATE ION, TYROSINE-PROTEIN PHOSPHATASE YIL113W | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2006-08-09 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Redox-mediated substrate recognition by Sdp1 defines a new group of tyrosine phosphatases.
Nature, 447, 2007
|
|
1MVU
 
 | |
2JBX
 
 | | Crystal Structure of the myxoma virus anti-apoptotic protein M11L | | Descriptor: | M11L PROTEIN | | Authors: | Kvansakul, M, Van Delft, M.F, Lee, E.F, Gulbis, J.M, Fairlie, W.D, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2006-12-14 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A Structural Viral Mimic of Prosurvival Bcl-2: A Pivotal Role for Sequestering Proapoptotic Bax and Bak.
Mol.Cell, 25, 2007
|
|
2JZH
 
 | | structure of IIB domain of the mannose transporter of E. coli | | Descriptor: | PTS system mannose-specific EIIAB component | | Authors: | Komlosh, M, Williams Jr, D.C. | | Deposit date: | 2008-01-08 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structures of Productive and Non-productive Complexes between the A and B Domains of the Cytoplasmic Subunit of the Mannose Transporter of the Escherichia coli Phosphotransferase System.
J.Biol.Chem., 283, 2008
|
|
4HMG
 
 | | REFINEMENT OF THE INFLUENZA VIRUS HEMAGGLUTININ BY SIMULATED ANNEALING | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, CHAIN HA1, ... | | Authors: | Weis, W.I, Bruenger, A.T, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1989-09-11 | | Release date: | 1991-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Refinement of the influenza virus hemagglutinin by simulated annealing.
J.Mol.Biol., 212, 1990
|
|
2F1G
 
 | | Cathepsin S in complex with non-covalent 2-(Benzoxazol-2-ylamino)-acetamide | | Descriptor: | Cathepsin S, GLYCEROL, N~2~-1,3-BENZOXAZOL-2-YL-3-CYCLOHEXYL-N-{2-[(4-METHOXYPHENYL)AMINO]ETHYL}-L-ALANINAMIDE | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S, Kulathila, R, Clark, K. | | Deposit date: | 2005-11-14 | | Release date: | 2006-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and evaluation of arylaminoethyl amides as noncovalent inhibitors of cathepsin S. Part 3: Heterocyclic P3.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2F5I
 
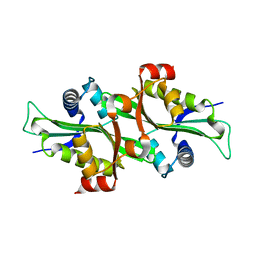 | |
2HXW
 
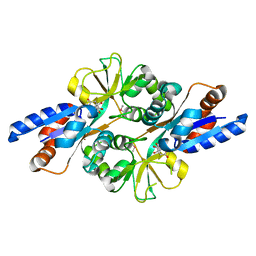 | | Crystal Structure of Peb3 from Campylobacter jejuni | | Descriptor: | CITRATE ANION, Major antigenic peptide PEB3 | | Authors: | Rangarajan, E.S, Bhatia, S, Watson, D.C, Munger, C, Cygler, M, Matte, A, Young, N.M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-08-04 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural context for protein N-glycosylation in bacteria: The structure of PEB3, an adhesin from Campylobacter jejuni.
Protein Sci., 16, 2007
|
|
2GJ0
 
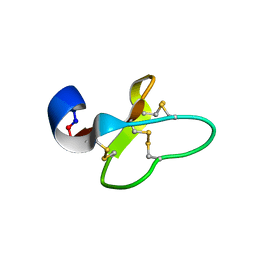 | | Cycloviolacin O14 | | Descriptor: | Cycloviolacin O14 | | Authors: | Ireland, D.C, Colgrave, M.L, Craik, D.J. | | Deposit date: | 2006-03-30 | | Release date: | 2006-04-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A novel suite of cyclotides from Viola odorata: sequence variation and the implications for structure, function and stability
Biochem.J., 400, 2006
|
|
2JM6
 
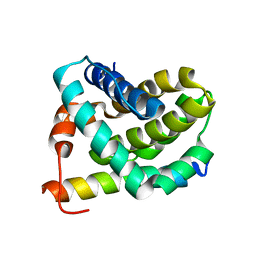 | | Solution structure of MCL-1 complexed with NOXAB | | Descriptor: | Myeloid cell leukemia-1 protein Mcl-1 homolog, Noxa | | Authors: | Czabotar, P.E, Lee, E.F, van Delft, M.F, Day, C.L, Smith, B.J, Huang, D.C.S, Fairlie, W.D, Hinds, M.G, Colman, P.M. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the degradation of Mcl-1 induced by BH3 domains
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2JBY
 
 | | A viral protein unexpectedly mimics the structure and function of pro- survival Bcl-2 | | Descriptor: | BCL-2 HOMOLOGOUS ANTAGONIST/KILLER 2, M11L PROTEIN, SODIUM ION | | Authors: | Kvansakul, M, Van Delft, M.F, Lee, E.F, Gulbis, J.M, Fairlie, W.D, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2006-12-14 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A structural viral mimic of prosurvival Bcl-2: a pivotal role for sequestering proapoptotic Bax and Bak.
Mol. Cell, 25, 2007
|
|
2KMG
 
 | | The structure of the KlcA and ArdB proteins show a novel fold and antirestriction activity against Type I DNA restriction systems in vivo but not in vitro | | Descriptor: | KlcA | | Authors: | Serfiotis-Mitsa, D, Herbert, A.P, Roberts, G.A, Soares, D.C, White, J.H, Blakely, G.W, Uhrin, D, Dryden, D.T.F. | | Deposit date: | 2009-07-28 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the KlcA and ArdB proteins reveals a novel fold and antirestriction activity against Type I DNA restriction systems in vivo but not in vitro
Nucleic Acids Res., 38, 2010
|
|
7UDK
 
 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 bound to LRPx4 peptide | | Descriptor: | 4xLRP, Designed helical repeat protein (DHR) RPB_LRP2_R4 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDL
 
 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 bound to PLPx6 peptide | | Descriptor: | 1,2-ETHANEDIOL, 6xPLP Peptide, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDN
 
 | |
7UDM
 
 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 in alternative conformation 1 (with peptide) | | Descriptor: | 6xPLP, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDO
 
 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 (proteolysis fragment?), forming pseudopolymeric filaments | | Descriptor: | 1,2-ETHANEDIOL, Designed helical repeat protein (DHR) RPB_LRP2_R4, PHOSPHATE ION | | Authors: | Redler, R.L, Chang, Y, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
