5H3O
 
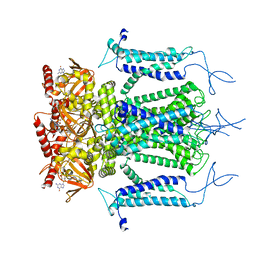 | | Structure of a eukaryotic cyclic nucleotide-gated channel | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel, SODIUM ION | | Authors: | Li, M, Zhou, X, Wang, S, Michailidis, I, Gong, Y, Su, D, Li, H, Li, X, Yang, J. | | Deposit date: | 2016-10-26 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of a eukaryotic cyclic-nucleotide-gated channel.
Nature, 542, 2017
|
|
5A3Z
 
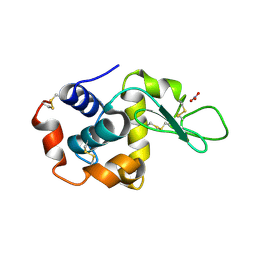 | | Structure of monoclinic Lysozyme obtained by multi crystal data collection | | Descriptor: | GLYCEROL, LYSOZYME, NITRATE ION | | Authors: | Zander, U, Bourenkov, G, Popov, A.N, de Sanctis, D, McCarthy, A.A, Svensson, O, Round, E.S, Gordeliy, V.I, Mueller-Dieckmann, C, Leonard, G.A. | | Deposit date: | 2015-06-04 | | Release date: | 2015-11-11 | | Last modified: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Meshandcollect: An Automated Multi-Crystal Data-Collection Workflow for Synchrotron Macromolecular Crystallography Beamlines.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7Q4B
 
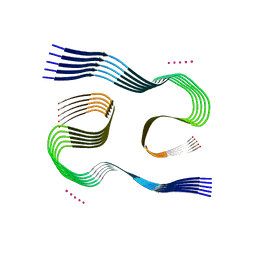 | | Type I beta-amyloid 42 Filaments from Human Brain | | Descriptor: | Amyloid-beta precursor protein, UNKNOWN ATOM OR ION | | Authors: | Yang, Y, Arseni, D, Zhang, W, Huang, M, Lovestam, S.K.A, Schweighauser, M, Kotecha, A, Murzin, A.G, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Garringer, H.J, Gelpi, E, Newell, K.L, Kovacs, G.G, Vidal, R, Ghetti, B, Falcon, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-10-30 | | Release date: | 2021-11-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structures of amyloid-beta 42 filaments from human brains.
Science, 375, 2022
|
|
5A45
 
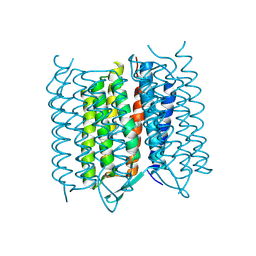 | | Structure of Bacteriorhodopsin obtained from 5um crystals by multi crystal data collection | | Descriptor: | BACTERIORHODOPSIN, RETINAL | | Authors: | Zander, U, Bourenkov, G, Popov, A.N, de Sanctis, D, McCarthy, A.A, Svensson, O, Round, E.S, Gordeliy, V.I, Mueller-Dieckmann, C, Leonard, G.A. | | Deposit date: | 2015-06-05 | | Release date: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Meshandcollect: An Automated Multi-Crystal Data-Collection Workflow for Synchrotron Macromolecular Crystallography Beamlines.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7PJA
 
 | | Crystal structure of YTHDC1 with compound PSI_DC1_002 | | Descriptor: | 6-methyl-3H-pyridin-2-one, GLYCEROL, SULFATE ION, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-08-23 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
1W5X
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | (2R,3R,4R,5R)-2,5-BIS[(2,3-DIFLUOROBENZYL)OXY]-3,4-DIHYDROXY-N,N'-BIS[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]HEXAN EDIAMIDE, POL POLYPROTEIN | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
7PJ7
 
 | |
5AFH
 
 | | alpha7-AChBP in complex with lobeline | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN, NEURONAL ACETYLCHOLINE RECEPTOR SUBUNIT ALPHA-7, ... | | Authors: | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3FPV
 
 | | Crystal Structure of HbpS | | Descriptor: | Extracellular haem-binding protein, FE (III) ION | | Authors: | Ortiz de Orue Lucana, D, Bogel, G, Zou, P, Groves, M.R. | | Deposit date: | 2009-01-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The oligomeric assembly of the novel haem-degrading protein HbpS is essential for interaction with its cognate two-component sensor kinase
J.Mol.Biol., 386, 2009
|
|
1W0U
 
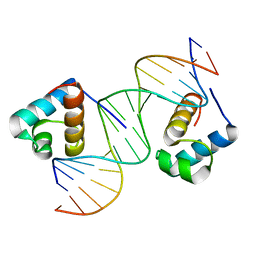 | | hTRF2 DNA-binding domain in complex with telomeric DNA. | | Descriptor: | 5'-D(*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP *CP*CP*CP*TP*AP*GP*A)-3', 5'-D(*TP*CP*TP*AP*GP*GP*GP*TP*TP*AP *GP*GP*GP*TP*TP*AP*G)-3', TELOMERIC REPEAT BINDING FACTOR 2 | | Authors: | Court, R.I, Chapman, L.M, Fairall, L, Rhodes, D. | | Deposit date: | 2004-06-11 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | How the Human Telomeric Proteins Trf1 and Trf2 Recognize Telomeric DNA: A View from High-Resolution Crystal Structures
Embo Rep., 6, 2005
|
|
5AJ3
 
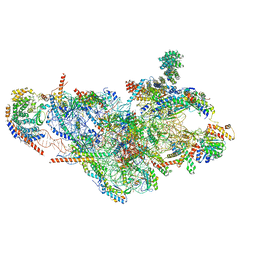 | | Structure of the small subunit of the mammalian mitoribosome | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MITORIBOSOMAL 12S RRNA, ... | | Authors: | Greber, B.J, Bieri, P, Leibundgut, M, Leitner, A, Aebersold, R, Boehringer, D, Ban, N. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-22 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome.
Science, 348, 2015
|
|
1VET
 
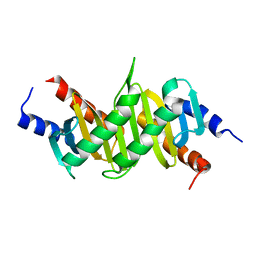 | | Crystal Structure of p14/MP1 at 1.9 A resolution | | Descriptor: | Late endosomal/lysosomal Mp1 interacting protein, Mitogen-activated protein kinase kinase 1 interacting protein 1 | | Authors: | Kurzbauer, R, Teis, D, Maurer-Stroh, S, Eisenhaber, F, Hekman, M, Bourenkov, G.P, Bartunik, H.D, Huber, L.A, Clausen, T. | | Deposit date: | 2004-04-05 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the p14/MP1 scaffolding complex: How a twin couple attaches mitogen- activated protein kinase signaling to late endosomes
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5AGT
 
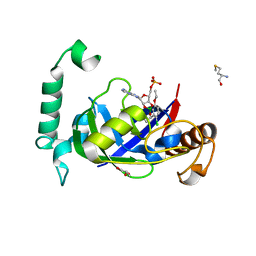 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct (S)-3-(Aminomethyl)-4-chloro-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | Descriptor: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, GLYCEROL, LEUCINE--TRNA LIGASE, ... | | Authors: | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5ACM
 
 | | Mcg immunoglobulin variable domain with methylene blue | | Descriptor: | 3,7-BIS(DIMETHYLAMINO)PHENOTHIAZIN-5-IUM, GLYCEROL, MCG, ... | | Authors: | Brumshtein, B, Esswein, S.R, Salwinski, L, Phillips, M.L, Ly, A.T, Cascio, D, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Inhibition by small-molecule ligands of formation of amyloid fibrils of an immunoglobulin light chain variable domain.
Elife, 4, 2015
|
|
5AEK
 
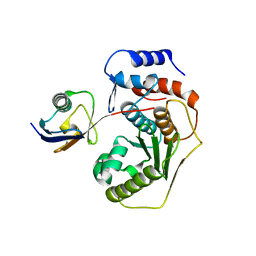 | | Crystal structure of the human SENP2 C548S in complex with the human SUMO1 K48M F66W | | Descriptor: | SENTRIN-SPECIFIC PROTEASE 2, SMALL UBIQUITIN-RELATED MODIFIER 1 | | Authors: | Gallego, P, Grana-Montes, R, Espargaro, A, Castillo, V, Torrent, J, Lange, R, Papaleo, E, Lindorff-Larsend, K, Ventura, S, Reverter, D. | | Deposit date: | 2014-12-23 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Stepping Back and Forward on Sumo Folding Evolution
To be Published
|
|
6QPH
 
 | | Dunaliella minimal PSI complex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Klaiman, D, Caspy, I, Nelson, N. | | Deposit date: | 2019-02-14 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a minimal photosystem I from the green alga Dunaliella salina.
Nat.Plants, 6, 2020
|
|
7PRZ
 
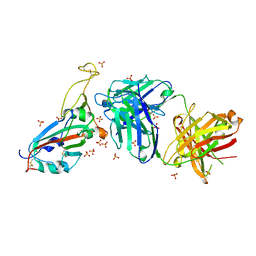 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with beta-22 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-22 Fab heavy chain, Beta-22 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
1W8U
 
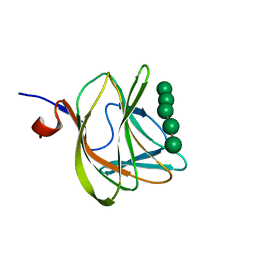 | | CBM29-2 mutant D83A complexed with mannohexaose: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON CATALYTIC PROTEIN 1, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-09-28 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
7PS2
 
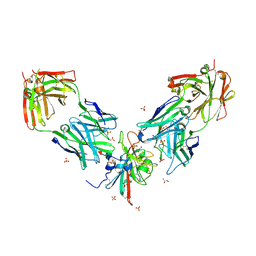 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-29 and Beta-53 Fabs | | Descriptor: | Beta-29 Fab heavy chain, Beta-29 Fab light chain, Beta-53 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
5AF5
 
 | | Structure of Lys33-linked triUb S.G. P 212121 | | Descriptor: | POLYUBIQUITIN-C | | Authors: | Michel, M.A, Elliott, P.R, Swatek, K.N, Simicek, M, Pruneda, J.N, Wagstaff, J.L, Freund, S.M.V, Komander, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Assembly and Specific Recognition of K29- and K33-Linked Polyubiquitin.
Mol.Cell, 58, 2015
|
|
7PS3
 
 | | Crystal structure of antibody Beta-32 Fab | | Descriptor: | Beta-32 heavy chain, Beta-32 light chain, CHLORIDE ION, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
1W9B
 
 | | S. alba myrosinase in complex with carba-glucotropaeolin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBA-GLUCOTROPAEOLIN, ... | | Authors: | Bourderioux, A, Lefoix, M, Gueyrard, D, Tatibouet, A, Cottaz, S, Arzt, S, Burmeister, W.P, Rollin, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The glucosinolate-myrosinase system. New insights into enzyme-substrate interactions by use of simplified inhibitors.
Org. Biomol. Chem., 3, 2005
|
|
5GVR
 
 | | Crystal structure of the DDX41 DEAD domain in an apo closed form | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Probable ATP-dependent RNA helicase DDX41 | | Authors: | Omura, H, Oikawa, D, Nakane, T, Kato, M, Ishii, R, Goto, Y, Suga, H, Ishitani, R, Tokunaga, F, Nureki, O. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide
Sci Rep, 6, 2016
|
|
3OY1
 
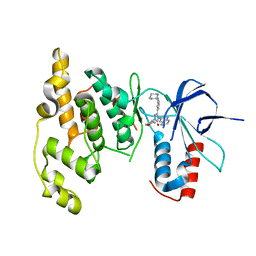 | | Highly Selective c-Jun N-Terminal Kinase (JNK) 2 and 3 Inhibitors with In Vitro CNS-like Pharmacokinetic Properties | | Descriptor: | 5-[2-(cyclohexylamino)pyridin-4-yl]-4-naphthalen-2-yl-2-(tetrahydro-2H-pyran-4-yl)-2,4-dihydro-3H-1,2,4-triazol-3-one, Mitogen-activated protein kinase 10 | | Authors: | Probst, G.D, Bowers, S, Sealy, J.M, Truong, A, Neitz, J, Hom, R.K, Galemmo Jr, R.A, Konradi, A.W, Sham, H.L, Quincy, D, Pan, H, Yao, N. | | Deposit date: | 2010-09-22 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Highly selective c-Jun N-terminal kinase (JNK) 2 and 3 inhibitors with in vitro CNS-like pharmacokinetic properties prevent neurodegeneration.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7PY2
 
 | | Structure of pathological TDP-43 filaments from ALS with FTLD | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Arseni, D, Hasegawa, H, Murzin, A.G, Kametani, F, Arai, M, Yoshida, M, Falcon, B. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structure of pathological TDP-43 filaments from ALS with FTLD.
Nature, 601, 2022
|
|
