7YX4
 
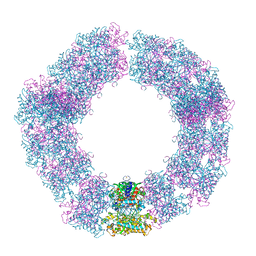 | | Structure of the Mimivirus genomic fibre in its compact 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
7YX3
 
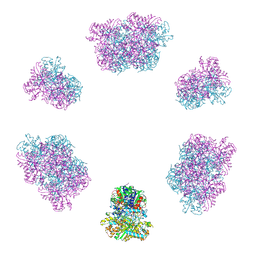 | | Structure of the Mimivirus genomic fibre in its compact 6-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30-nm diameter helical protein shield.
Elife, 11, 2022
|
|
3E3M
 
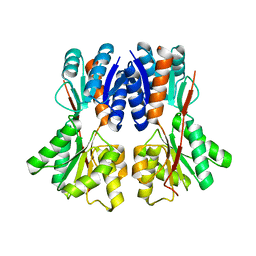 | | Crystal structure of a LacI family transcriptional regulator from Silicibacter pomeroyi | | Descriptor: | Transcriptional regulator, LacI family | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-07 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a LacI family transcriptional regulator from Silicibacter pomeroyi
To be Published
|
|
3E9G
 
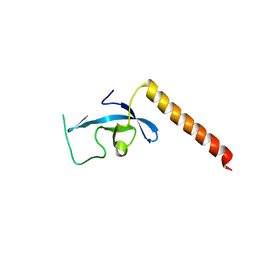 | | Crystal structure long-form (residue1-124) of Eaf3 chromo domain | | Descriptor: | Chromatin modification-related protein EAF3 | | Authors: | Sun, B, Hong, J, Zhang, P, Lin, D, Ding, J. | | Deposit date: | 2008-08-22 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis of the Interaction of Saccharomyces cerevisiae Eaf3 Chromo Domain with Methylated H3K36
J.Biol.Chem., 283, 2008
|
|
3EAQ
 
 | |
7ZC8
 
 | |
3EAY
 
 | | Crystal structure of the human SENP7 catalytic domain | | Descriptor: | SULFATE ION, Sentrin-specific protease 7 | | Authors: | Lima, C.D, Reverter, D. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Human SENP7 Catalytic Domain and Poly-SUMO Deconjugation Activities for SENP6 and SENP7.
J.Biol.Chem., 283, 2008
|
|
3PU2
 
 | | Crystal Structure of the Q3J4M4_RHOS4 protein from Rhodobacter sphaeroides. Northeast Structural Genomics Consortium Target RhR263. | | Descriptor: | uncharacterized protein | | Authors: | Vorobiev, S, Chen, Y, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-15 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Crystal Structure of the Q3J4M4_RHOS4 protein from Rhodobacter sphaeroides.
To be Published
|
|
2IJ4
 
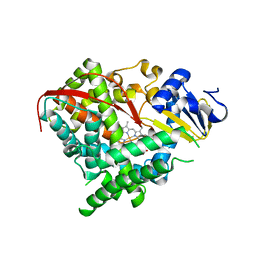 | | Structure of the A264K mutant of cytochrome P450 BM3 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 BM3 | | Authors: | Toogood, H.S, Leys, D. | | Deposit date: | 2006-09-29 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and spectroscopic characterization of P450 BM3 mutants with unprecedented P450 heme iron ligand sets. New heme ligation states influence conformational equilibria in P450 BM3.
J.Biol.Chem., 282, 2007
|
|
3VZ3
 
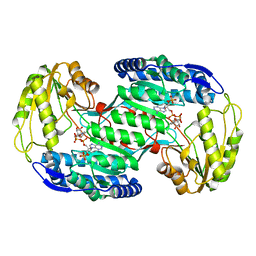 | | Structural insights into substrate and cofactor selection by sp2771 | | Descriptor: | 4-oxobutanoic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Succinate-semialdehyde dehydrogenase | | Authors: | Yuan, Y.A, Yuan, Z, Yin, B, Wei, D. | | Deposit date: | 2012-10-09 | | Release date: | 2013-07-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis for cofactor and substrate selection by cyanobacterium succinic semialdehyde dehydrogenase
J.Struct.Biol., 182, 2013
|
|
3W0R
 
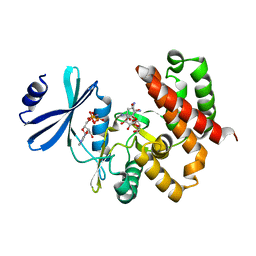 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia (N202A), ternary complex with AMP-PNP and hygromycin B | | Descriptor: | HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
5TIC
 
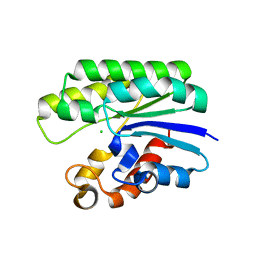 | | X-ray structure of wild-type E. coli Acyl-CoA thioesterase I at pH 5 | | Descriptor: | Acyl-CoA thioesterase I, CHLORIDE ION | | Authors: | Thoden, J.B, Holden, H.M, Grisewood, M.J, Hernandez Lozada, N.J, Gifford, N.P, Mendez-Perez, D, Schoenberger, H.A, Allan, M.F, Pfleger, B.F, Marines, C.D. | | Deposit date: | 2016-10-02 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Computational Redesign of Acyl-ACP Thioesterase with Improved Selectivity toward Medium-Chain-Length Fatty Acids.
ACS Catal, 7, 2017
|
|
3EU5
 
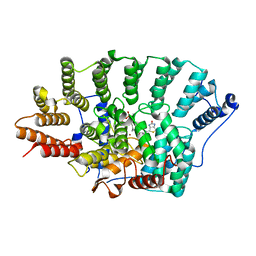 | | Crystal structure of FTase(ALPHA-subunit; BETA-subunit DELTA C10) in complex with BiotinGPP | | Descriptor: | (2E,6E)-3,7-dimethyl-8-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)octa-2,6-dien-1-yl trihydrogen diphosphate, Protein farnesyltransferase subunit beta, Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha, ... | | Authors: | Guo, Z, Nguyen, U.T.T, Delon, C, Bon, R.S, Blankenfeldt, W, Goody, R.S, Waldmann, H, Wolters, D, Alexandrov, K. | | Deposit date: | 2008-10-09 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the eukaryotic prenylome by isoprenoid affinity tagging
Nat.Chem.Biol., 5, 2009
|
|
7Z1K
 
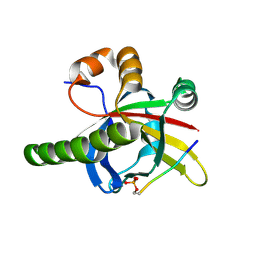 | | Crystal structure of the SPOC domain of human SHARP (SPEN) in complex with RNA polymerase II CTD heptapeptide phosphorylated on Ser5 | | Descriptor: | Msx2-interacting protein, SER-TYR-SER-PRO-THR-SEP | | Authors: | Appel, L, Grishkovskaya, I, Slade, D, Djinovic-Carugo, K. | | Deposit date: | 2022-02-24 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The SPOC domain is a phosphoserine binding module that bridges transcription machinery with co- and post-transcriptional regulators.
Nat Commun, 14, 2023
|
|
7Z27
 
 | |
2IRV
 
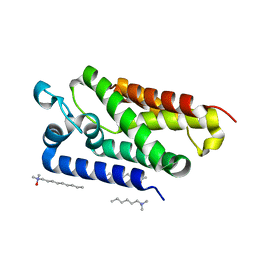 | | Crystal structure of GlpG, a rhomboid intramembrane serine protease | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, DODECYL-BETA-D-MALTOSIDE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Bibi, E, Fass, D, Ben-Shem, A. | | Deposit date: | 2006-10-16 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for intramembrane proteolysis by rhomboid serine proteases.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2IUR
 
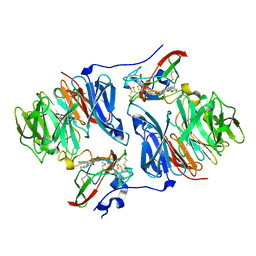 | |
3VBF
 
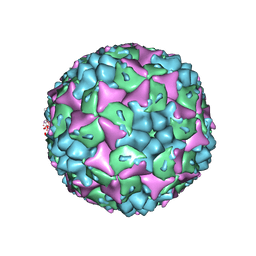 | | Crystal structure of formaldehyde treated human Enterovirus 71 (space group I23) | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Genome Polyprotein, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5TID
 
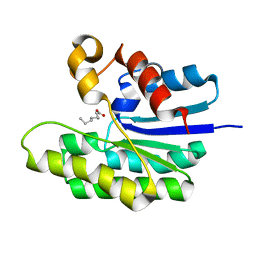 | | X-ray structure of acyl-CoA thioesterase I, TesA, mutant M141L/Y145K/L146K at pH 5 in complex with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Thoden, J.B, Holden, H.M, Grisewood, M.J, Hernandez Lozada, N.J, Gifford, N.P, Mendez-Perez, D, Schoenberger, H.A, Allan, M.F, Pfleger, B.F, Marines, C.D. | | Deposit date: | 2016-10-02 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Computational Redesign of Acyl-ACP Thioesterase with Improved Selectivity toward Medium-Chain-Length Fatty Acids.
ACS Catal, 7, 2017
|
|
2J14
 
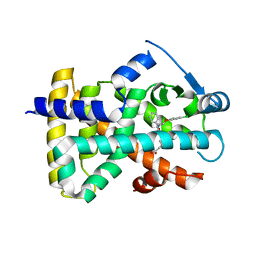 | | 3,4,5-Trisubstituted Isoxazoles as Novel PPARdelta Agonists: Part2 | | Descriptor: | (3-{4-[2-(2,4-DICHLORO-PHENOXY)-ETHYLCARBAMOYL]-5-PHENYL-ISOXAZOL-3-YL}-PHENYL)-ACETIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR DELTA | | Authors: | Epple, R, Azimioara, M, Russo, R, Xie, Y, Wang, X, Cow, C, Wityak, J, Karanewsky, D, Bursulaya, B, Kreusch, A, Tuntland, T, Gerken, A, Iskandar, M, Saez, E, Seidel, H.M, Tian, S.S. | | Deposit date: | 2006-08-08 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3,4,5-Trisubstituted Isoxazoles as Novel Ppardelta Agonists. Part 2
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3PPF
 
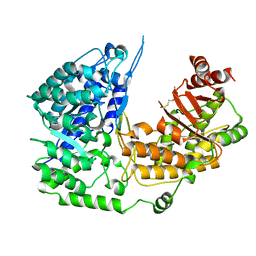 | | Crystal structure of the Candida albicans methionine synthase by surface entropy reduction, alanine variant without zinc | | Descriptor: | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | | Authors: | Ubhi, D, Kavanagh, K, Monzingo, A.F, Robertus, J.D. | | Deposit date: | 2010-11-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Candida albicans methionine synthase determined by employing surface residue mutagenesis.
Arch.Biochem.Biophys., 513, 2011
|
|
7YL7
 
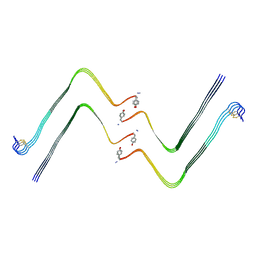 | | Structure of hIAPP-TF-type3 | | Descriptor: | Islet amyloid polypeptide | | Authors: | Li, D, Zhang, X. | | Deposit date: | 2022-07-25 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A new polymorphism of human amylin fibrils with similar protofilaments and a conserved core.
Iscience, 25, 2022
|
|
7YM0
 
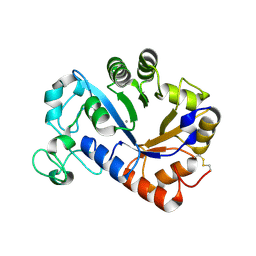 | | Lysoplasmalogen-specific phospholipase D (LyPls-PLD) with Ca2+ | | Descriptor: | CALCIUM ION, Lysoplasmalogenase | | Authors: | Yasutake, Y, Sakasegawa, S, Sugimori, D, Murayama, K. | | Deposit date: | 2022-07-27 | | Release date: | 2023-01-04 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural basis for the substrate specificity switching of lysoplasmalogen-specific phospholipase D from Thermocrispum sp. RD004668.
Biosci.Biotechnol.Biochem., 87, 2022
|
|
7YYO
 
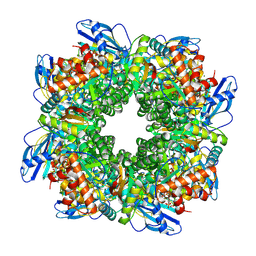 | | Cryo-EM structure of an a-carboxysome RuBisCO enzyme at 2.9 A resolution | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Mann, D, Evans, S.L, Bergeron, J.R.C. | | Deposit date: | 2022-02-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Single-particle cryo-EM analysis of the shell architecture and internal organization of an intact alpha-carboxysome.
Structure, 31, 2023
|
|
5U4W
 
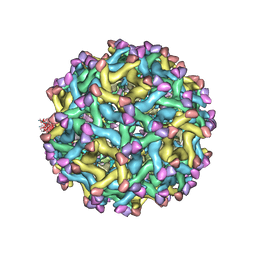 | | Cryo-EM Structure of Immature Zika Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, E protein, M protein, ... | | Authors: | Mangala Prasad, V, Miller, A.S, Klose, T, Sirohi, D, Buda, G, Jiang, W, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Structure of the immature Zika virus at 9 angstrom resolution.
Nat. Struct. Mol. Biol., 24, 2017
|
|
