4UUC
 
 | | Crystal structure of human ASB11 ankyrin repeat domain | | Descriptor: | ANKYRIN REPEAT AND SOCS BOX PROTEIN 11 | | Authors: | Pinkas, D.M, Sanvitale, C, Kragh Nielsen, T, Guo, K, Sorrell, F, Berridge, G, Ayinampudi, V, Wang, D, Newman, J.A, Tallant, C, Chaikuad, A, Canning, P, Kopec, J, Krojer, T, Vollmar, M, Allerston, C.K, Chalk, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Bullock, A. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Asb11 Ankyrin Repeat Domain
To be Published
|
|
4UUJ
 
 | | POTASSIUM CHANNEL KCSA-FAB WITH TETRAHEXYLAMMONIUM | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, COBALT (II) ION, ... | | Authors: | Lenaeus, M.J, Burdette, D, Wagner, T, Focia, P.J, Gross, A. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Kcsa in Complex with Symmetrical Quaternary Ammonium Compounds Reveal a Hydrophobic Binding Site.
Biochemistry, 53, 2014
|
|
6NZX
 
 | | Hadesarchaea YNP_N21 cytochrome b5 domain protein (KUO41884.1) | | Descriptor: | Cytochrome B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Teakel, S.L, Marama, M.S, Aragao, D, Forwood, J.K, Cahill, M.A. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hadesarchaea YNP_N21 cytochrome b5 domain protein (KUO41884.1)
To Be Published
|
|
1DDN
 
 | | DIPHTHERIA TOX REPRESSOR (C102D MUTANT)/TOX DNA OPERATOR COMPLEX | | Descriptor: | 33 BASE DNA CONTAINING TOXIN OPERATOR, DIPHTHERIA TOX REPRESSOR, NICKEL (II) ION | | Authors: | White, A, Ding, X, Vanderspek, J.C, Murphy, J.R, Ringe, D. | | Deposit date: | 1998-06-23 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the metal-ion-activated diphtheria toxin repressor/tox operator complex.
Nature, 394, 1998
|
|
7M31
 
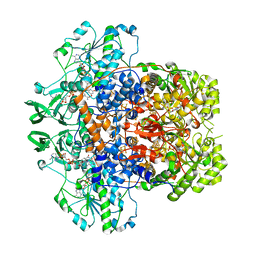 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Thymine and NADPH Anaerobically | | Descriptor: | Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Butrin, A, Beaupre, B, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Perturbing the Movement of Hydrogens to Delineate and Assign Events in the Reductive Activation and Turnover of Porcine Dihydropyrimidine Dehydrogenase.
Biochemistry, 60, 2021
|
|
7M32
 
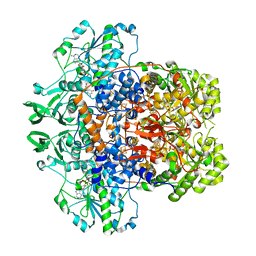 | | Dihydropyrimidine Dehydrogenase (DPD) C671A Mutant Soaked with Uracil and NADPH Anaerobically | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ALANINE, Dihydropyrimidine dehydrogenase [NADP(+)], ... | | Authors: | Butrin, A, Beaupre, B, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Perturbing the Movement of Hydrogens to Delineate and Assign Events in the Reductive Activation and Turnover of Porcine Dihydropyrimidine Dehydrogenase.
Biochemistry, 60, 2021
|
|
4V39
 
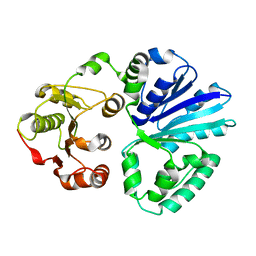 | | Apo-structure of alpha2,3-sialyltransferase variant 2 from Pasteurella dagmatis | | Descriptor: | SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
4V0W
 
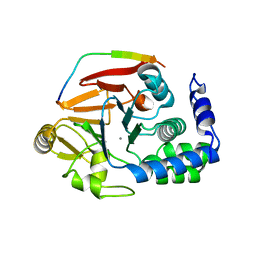 | | The crystal structure of mouse PP1G in complex with truncated human PPP1R15B (631-669) | | Descriptor: | MANGANESE (II) ION, PROTEIN PHOSPHATASE 1 REGULATORY SUBUNIT 15B, SERINE/THREONINE-PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT | | Authors: | Chen, R, Yan, Y, Casado, A.C, Ron, D, Read, R.J. | | Deposit date: | 2014-09-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | G-actin provides substrate-specificity to eukaryotic initiation factor 2 alpha holophosphatases.
Elife, 4, 2015
|
|
4UWL
 
 | | Discovery of (2S)-8-((3R)-3-Methylmorpholin-4-yl)-1-(3-methyl-2-oxo- butyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido(1,2-a)pyrimidin-6- one: a Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors | | Descriptor: | (8S)-2-[(3R)-3-methylmorpholin-4-yl]-9-(3-methyl-2-oxobutyl)-8-(trifluoromethyl)-6,7,8,9-tetrahydro-4H-pyrimido[1,2-a]pyrimidin-4-one, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT TYPE 3, SULFATE ION | | Authors: | Pasquier, B, El-Ahmad, Y, Filoche-Romme, B, Dureuil, C, Fassy, F, Abecassis, P.Y, Mathieu, M, Bertrand, T, Benard, T, Barriere, C, ElBatti, S, Letallec, J.P, Sonnefraud, V, Brollo, M, Delbarre, L, Loyau, V, Pilorge, F, Bertin, L, Richepin, P, Arigon, J, Labrosse, J.R, Clement, J, Durand, F, Combet, R, Perraut, P, Leroy, V, Gay, F, Lefrancois, D, Bretin, F, Marquette, J.P, Michot, N, Caron, A, Castell, C, Schio, L, McCort, G, Goulaouic, H, Garcia-Echeverria, C, Ronan, B. | | Deposit date: | 2014-08-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-Yl]-1-(3-Methyl-2-Oxo-Butyl)-2-(Trifluoromethyl)-3,4-Dihydro-2H-Pyrimido[1,2-A]Pyrimidin-6-One: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors.
J.Med.Chem., 58, 2015
|
|
2YPA
 
 | | Structure of the SCL:E47:LMO2:LDB1 complex bound to DNA | | Descriptor: | EBOX FORWARD, EBOX REVERSE, LIM DOMAIN-BINDING PROTEIN 1, ... | | Authors: | El Omari, K, Hoosdally, S.J, Tuladhar, K, Karia, D, Ponsele, E, Platonova, O, Vyas, P, Patient, R, Porcher, C, Mancini, E.J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Lmo2-Driven Recruitment of the Scl:E47bHLH Heterodimer to Hematopoietic-Specific Transcriptional Targets.
Cell Rep., 4, 2013
|
|
4UWK
 
 | | Discovery of (2S)-8-((3R)-3-Methylmorpholin-4-yl)-1-(3-methyl-2-oxo- butyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido(1,2-a)pyrimidin-6- one: a Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors | | Descriptor: | (2S)-1-[(5-chloro-2-thienyl)methyl]-8-[(3R,5R)-3,5-dimethylmorpholin-4-yl]-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido[1,2-a]pyrimidin-6-one, GLYCEROL, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT TYPE 3, ... | | Authors: | Pasquier, B, El-Ahmad, Y, Filoche-Romme, B, Dureuil, C, Fassy, F, Abecassis, P.Y, Mathieu, M, Bertrand, T, Benard, T, Barriere, C, ElBatti, S, Letallec, J.P, Sonnefraud, V, Brollo, M, Delbarre, L, Loyau, V, Pilorge, F, Bertin, L, Richepin, P, Arigon, J, Labrosse, J.R, Clement, J, Durand, F, Combet, R, Perraut, P, Leroy, V, Gay, F, Lefrancois, D, Bretin, F, Marquette, J.P, Michot, N, Caron, A, Castell, C, Schio, L, McCort, G, Goulaouic, H, Garcia-Echeverria, C, Ronan, B. | | Deposit date: | 2014-08-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-Yl]-1-(3-Methyl-2-Oxo-Butyl)-2-(Trifluoromethyl)-3,4-Dihydro-2H-Pyrimido[1,2-A]Pyrimidin-6-One: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors.
J.Med.Chem., 58, 2015
|
|
4UWH
 
 | | Discovery of (2S)-8-((3R)-3-Methylmorpholin-4-yl)-1-(3-methyl-2-oxo- butyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido(1,2-a)pyrimidin-6- one: a Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors | | Descriptor: | (8S)-9-[(2R)-2-hydroxy-2-phenylethyl]-2-(morpholin-4-yl)-8-(trifluoromethyl)-6,7,8,9-tetrahydro-4H-pyrimido[1,2-a]pyrimidin-4-one, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT TYPE 3, SODIUM ION | | Authors: | Pasquier, B, El-Ahmad, Y, Filoche-Romme, B, Dureuil, C, Fassy, F, Abecassis, P.Y, Mathieu, M, Bertrand, T, Benard, T, Barriere, C, ElBatti, S, Letallec, J.P, Sonnefraud, V, Brollo, M, Delbarre, L, Loyau, V, Pilorge, F, Bertin, L, Richepin, P, Arigon, J, Labrosse, J.R, Clement, J, Durand, F, Combet, R, Perraut, P, Leroy, V, Gay, F, Lefrancois, D, Bretin, F, Marquette, J.P, Michot, N, Caron, A, Castell, C, Schio, L, McCort, G, Goulaouic, H, Garcia-Echeverria, C, Ronan, B. | | Deposit date: | 2014-08-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-Yl]-1-(3-Methyl-2-Oxo-Butyl)-2-(Trifluoromethyl)-3,4-Dihydro-2H-Pyrimido[1,2-A]Pyrimidin-6-One: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors.
J.Med.Chem., 58, 2015
|
|
4UXH
 
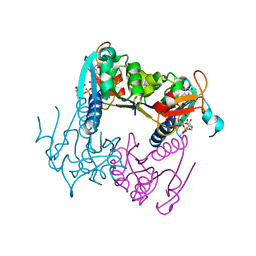 | | Leishmania major Thymidine Kinase in complex with AP5dT | | Descriptor: | P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, THYMIDINE KINASE, ZINC ION | | Authors: | Timm, J, Bosch-Navarrete, C, Recio, E, Nettleship, J.E, Rada, H, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2014-08-22 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Kinetic Characterization of Thymidine Kinase from Leishmania Major.
Plos Negl Trop Dis, 9, 2015
|
|
2YMZ
 
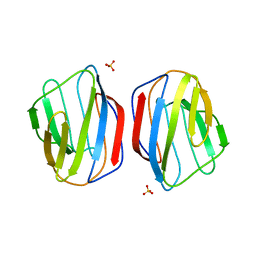 | | Crystal structure of chicken Galectin 2 | | Descriptor: | GALECTIN 2, SULFATE ION, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Fernandez, I.S, Ruiz, F.M, Solis, D, Gabius, H.-J, Romero, A. | | Deposit date: | 2012-10-11 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fine-Tuning of Prototype Chicken Galectins: Structure of Cg-2 and Structure-Activity Correlations
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2YQS
 
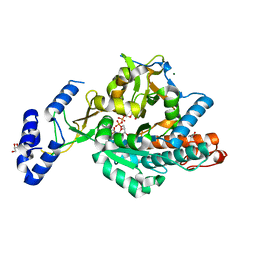 | | Crystal structure of uridine-diphospho-N-acetylglucosamine pyrophosphorylase from Candida albicans, in the product-binding form | | Descriptor: | GLYCEROL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Miki, K, Maruyama, D, Nishitani, Y, Nonaka, T, Kita, A. | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Uridine-diphospho-N-acetylglucosamine Pyrophosphorylase from Candida albicans and Catalytic Reaction Mechanism
J.Biol.Chem., 282, 2007
|
|
4UYI
 
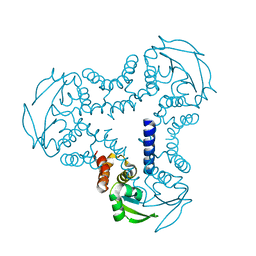 | | Crystal structure of the BTB domain of human SLX4 (BTBD12) | | Descriptor: | STRUCTURE-SPECIFIC ENDONUCLEASE SUBUNIT SLX4 | | Authors: | Pinkas, D.M, Sanvitale, C.E, Strain-Damerell, C, Fairhead, M, Wang, D, Tallant, C, Cooper, C.D.O, Sorrell, F.J, Kopec, J, Chaikuad, A, Fitzpatrick, F, Pike, A.C.W, Hozjan, V, Ying, Z, Roos, A.K, Savitsky, P, Bradley, A, Nowak, R, Filippakopoulos, P, Krojer, T, Burgess-Brown, N.A, Marsden, B.D, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-09-01 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the Btb Domain of Human Slx4 (Btbd12)
To be Published
|
|
1STC
 
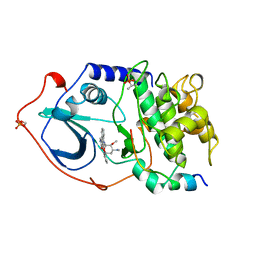 | | CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH STAUROSPORINE | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, PROTEIN KINASE INHIBITOR, STAUROSPORINE | | Authors: | Prade, L, Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1997-10-10 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Staurosporine-induced conformational changes of cAMP-dependent protein kinase catalytic subunit explain inhibitory potential.
Structure, 5, 1997
|
|
1DFN
 
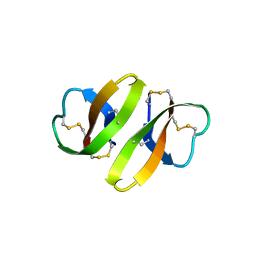 | | CRYSTAL STRUCTURE OF DEFENSIN HNP-3, AN AMPHIPHILIC DIMER: MECHANISMS OF MEMBRANE PERMEABILIZATION | | Descriptor: | DEFENSIN HNP-3 | | Authors: | Hill, C.P, Yee, J, Selsted, M.E, Eisenberg, D. | | Deposit date: | 1991-01-18 | | Release date: | 1992-07-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of defensin HNP-3, an amphiphilic dimer: mechanisms of membrane permeabilization.
Science, 251, 1991
|
|
3EHJ
 
 | |
6O3C
 
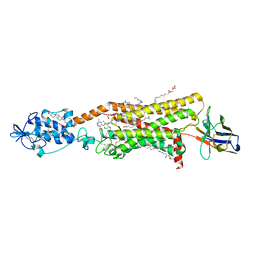 | | Crystal structure of active Smoothened bound to SAG21k, cholesterol, and NbSmo8 | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-chloro-4,7-difluoro-N-{[2-methoxy-5-(pyridin-4-yl)phenyl]methyl}-N-[trans-4-(methylamino)cyclohexyl]-1-benzothiophene-2-carboxamide, ... | | Authors: | Deshpande, I.S, Liang, J, Hedeen, D, Roberts, K.J, Zhang, Y, Ha, B, Latorraca, N.R, Faust, B, Dror, R.O, Beachy, P.A, Myers, B.R, Manglik, A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Smoothened stimulation by membrane sterols drives Hedgehog pathway activity.
Nature, 571, 2019
|
|
4V1X
 
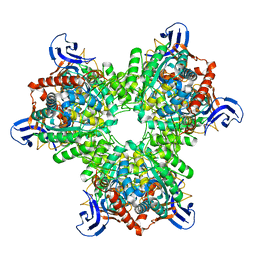 | | The structure of the hexameric atrazine chlorohydrolase, AtzA | | Descriptor: | ATRAZINE CHLOROHYDROLASE, DI(HYDROXYETHYL)ETHER, FE (III) ION | | Authors: | Peat, T.S, Newman, J, Balotra, S, Lucent, D, Warden, A.C, Scott, C. | | Deposit date: | 2014-10-04 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Hexameric Atrazine Chlorohydrolase Atza.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8GQ3
 
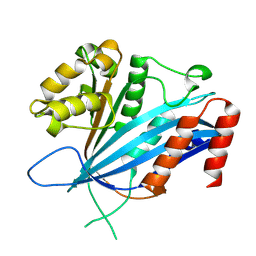 | | RTT109 mutant from Candida albicans | | Descriptor: | Histone acetyltransferase RTT109 | | Authors: | Chen, Y.J, Su, D. | | Deposit date: | 2022-08-28 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Crystal structure of RTT109 mutant-Y183A from Candida albicans
To Be Published
|
|
4UZM
 
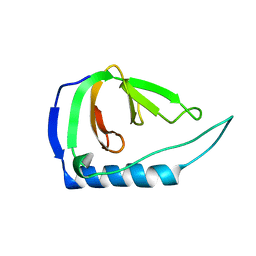 | |
1TCH
 
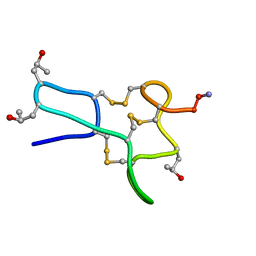 | |
4WH7
 
 | | Structure of the CDC25B Phosphatase Catalytic Domain with Bound Ligand | | Descriptor: | 2-fluoro-4-hydroxybenzonitrile, GLYCEROL, M-phase inducer phosphatase 2, ... | | Authors: | Lund, G.L, Dudkin, S, Borkin, D, Ni, W, Grembecka, J, Cierpicki, T. | | Deposit date: | 2014-09-20 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Inhibition of CDC25B Phosphatase Through Disruption of Protein-Protein Interaction.
Acs Chem.Biol., 10, 2015
|
|
