8CXN
 
 | |
8CY9
 
 | |
6WVT
 
 | | Structural basis of alphaE-catenin - F-actin catch bond behavior | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Xu, X.P, Pokutta, S, Torres, M, Swift, M.F, Hanein, D, Volkmann, N, Weis, W.I. | | Deposit date: | 2020-05-06 | | Release date: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis of alpha E-catenin-F-actin catch bond behavior.
Elife, 9, 2020
|
|
8CY7
 
 | |
6TIH
 
 | |
6WGW
 
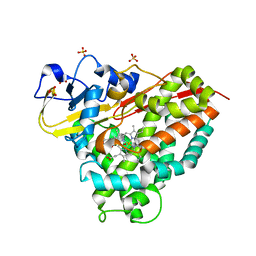 | | CYP101D1 D259E Hydroxycamphor bound | | Descriptor: | 5-EXO-HYDROXYCAMPHOR, CAMPHOR, Cytochrome P450 101D1, ... | | Authors: | Amaya, J.A, Poulos, T.L, Batabyal, D. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Proton Relay Network in the Bacterial P450s: CYP101A1 and CYP101D1.
Biochemistry, 59, 2020
|
|
3H6P
 
 | | Crystal structure of Rv3019c-Rv3020c from Mycobacterium tuberculosis | | Descriptor: | ESAT-6 LIKE PROTEIN ESXS, ESAT-6-like protein esxR, GLYCEROL | | Authors: | Chan, S, Arbing, M, Phan, T, Kaufmann, M, Cascio, D, Eisenberg, D, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2009-04-23 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of Rv3019c-Rv3020c from Mycobacterium tuberculosis
To be Published
|
|
6WNV
 
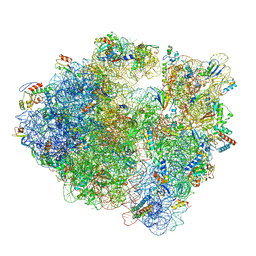 | | 70S ribosome without free 5S rRNA and with a perturbed PTC | | Descriptor: | 16S ribosomal RNA, 23s-5s joint ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Korostelev, A.A, Mankin, A.S, Huang, S, Aleksashin, N.A, Klepacki, D, Reier, K, Kefi, A, Szal, A, Remme, J, Jaeger, L, Vazquez-Laslop, N. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ribosome engineering reveals the importance of 5S rRNA autonomy for ribosome assembly.
Nat Commun, 11, 2020
|
|
6WI5
 
 | |
6TM6
 
 | | MUC2 CysD1 domain | | Descriptor: | CALCIUM ION, Mucin-2 | | Authors: | Khmelnitsky, L, Fass, D. | | Deposit date: | 2019-12-03 | | Release date: | 2020-02-19 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Assembly Mechanism of Mucin and von Willebrand Factor Polymers.
Cell, 183, 2020
|
|
6T8V
 
 | | Complement factor B in complex with (S)-5,7-Dimethyl-4-((2-phenylpiperidin-1-yl)methyl)-1H-indole | | Descriptor: | 4-[(2~{S})-1-[(5,7-dimethyl-1~{H}-indol-4-yl)methyl]piperidin-2-yl]benzoic acid, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
3H98
 
 | | Crystal structure of HCV NS5b 1b with (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyrimidine derivative | | Descriptor: | GLYCEROL, N-{3-[5-hydroxy-8-(3-methylbutyl)-7-oxo-7,8-dihydroimidazo[1,2-a]pyrimidin-6-yl]-1,1-dioxido-4H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Wang, G, Lei, H, Wang, X, Das, D, Mackinnon, C, Montalbetti, C.A.G, Mears, R, Gai, X, Bailey, S, Ruhrmund, D, Hooi, L, Misialek, S, Rajagopalan, R, Cheng, R.K.Y, Barker, J.L, Felicetti, B, Stoycheva, A, Buckman, B, Kossen, K, Seiwert, S, Beigelmana, L. | | Deposit date: | 2009-04-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HCV NS5B polymerase inhibitors 2: Synthesis and in vitro activity of (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyridine and azolo[1,5-a]pyrimidine derivatives.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8D8O
 
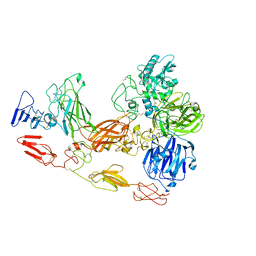 | | Cryo-EM structure of substrate unbound PAPP-A | | Descriptor: | Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-06-08 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
1E0V
 
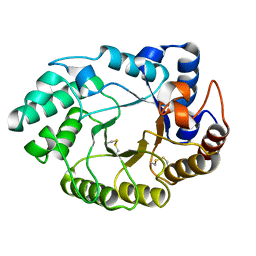 | | Xylanase 10A from Sreptomyces lividans. cellobiosyl-enzyme intermediate at 1.7 A | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
6TTG
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with LMD62 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-(2-morpholin-4-ylethoxy)-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, ... | | Authors: | Welin, M, Kimbung, R, Focht, D. | | Deposit date: | 2019-12-27 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | New dual ATP-competitive inhibitors of bacterial DNA gyrase and topoisomerase IV active against ESKAPE pathogens.
Eur.J.Med.Chem., 213, 2021
|
|
6WH8
 
 | | The structure of NTMT1 in complex with compound BM-30 | | Descriptor: | 4HP-PRO-LYS-ARG-NH2, BM-30, N-terminal Xaa-Pro-Lys N-methyltransferase 1, ... | | Authors: | Noinaj, N, Chen, D, Huang, R. | | Deposit date: | 2020-04-07 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | Selective Peptidomimetic Inhibitors of NTMT1/2: Rational Design, Synthesis, Characterization, and Crystallographic Studies.
J.Med.Chem., 63, 2020
|
|
8A85
 
 | |
6WPL
 
 | | Structure of Cytochrome P450tcu | | Descriptor: | 5-EXO-HYDROXYCAMPHOR, Cytochrome P-450cam, subunit of camphor 5-monooxygenase system, ... | | Authors: | Murarka, V.C, Batabyal, D, Amaya, J.A, Sevrioukova, I.F, Poulos, T.L. | | Deposit date: | 2020-04-27 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Unexpected Differences between Two Closely Related Bacterial P450 Camphor Monooxygenases.
Biochemistry, 59, 2020
|
|
6WPQ
 
 | | GNYNVF from hnRNPA2-low complexity domain segment, residues 286-291, D290V variant | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A2 | | Authors: | Lu, J, Cao, Q, Hughes, M.P, Sawaya, M.R, Boyer, D.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | CryoEM structure of the low-complexity domain of hnRNPA2 and its conversion to pathogenic amyloid.
Nat Commun, 11, 2020
|
|
6TIJ
 
 | |
7LL8
 
 | | D-Protein RFX-V1 Bound to the VEGFR1 Domain 2 Site on VEGF-A | | Descriptor: | Isoform L-VEGF189 of Vascular endothelial growth factor A, RFX-V1 | | Authors: | Marinec, P.S, Landgraf, K.E, Uppalapati, M, Chen, G, Xie, D, Jiang, Q, Zhao, Y, Petriello, A, Deshayes, K, Kent, S.B.H, Ault-Riche, D, Sidhu, S.S. | | Deposit date: | 2021-02-03 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A Non-immunogenic Bivalent d-Protein Potently Inhibits Retinal Vascularization and Tumor Growth.
Acs Chem.Biol., 16, 2021
|
|
7LL9
 
 | | D-Protein RFX-V2 Bound to the VEGFR1 Domain 3 Site on VEGF-A | | Descriptor: | Isoform L-VEGF189 of Vascular endothelial growth factor A, RFX-V2 | | Authors: | Marinec, P.S, Landgraf, K.E, Uppalapati, M, Chen, G, Xie, D, Jiang, Q, Zhao, Y, Petriello, A, Deshayes, K, Kent, S.B.H, Ault-Riche, D, Sidhu, S.S. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Non-immunogenic Bivalent d-Protein Potently Inhibits Retinal Vascularization and Tumor Growth.
Acs Chem.Biol., 16, 2021
|
|
7ZLW
 
 | | NME1 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nucleoside diphosphate kinase A | | Authors: | Garcia-Saez, I, Iuso, D, Khochbin, S, Petosa, C. | | Deposit date: | 2022-04-15 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nucleoside diphosphate kinases 1 and 2 regulate a protective liver response to a high-fat diet.
Sci Adv, 9, 2023
|
|
6WX9
 
 | | SOX2 bound to Importin-alpha 5 | | Descriptor: | Importin subunit alpha-5, Transcription factor SOX-2 | | Authors: | Bikshapathi, J, Stewart, M, Forwood, J.K, Aragao, D, Roman, N. | | Deposit date: | 2020-05-10 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for nuclear import selectivity of pioneer transcription factor SOX2.
Nat Commun, 12, 2021
|
|
7ZL8
 
 | | NME1 in complex with succinyl-CoA | | Descriptor: | Nucleoside diphosphate kinase A, SUCCINYL-COENZYME A | | Authors: | Garcia-Saez, I, Iuso, D, Khochbin, S, Petosa, C. | | Deposit date: | 2022-04-14 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Nucleoside diphosphate kinases 1 and 2 regulate a protective liver response to a high-fat diet.
Sci Adv, 9, 2023
|
|
