7WJU
 
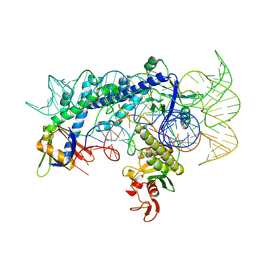 | | Cryo-EM structure of the AsCas12f1-sgRNAv1-dsDNA ternary complex | | Descriptor: | Non-target strand, OrfB_Zn_ribbon domain-containing protein, Target strand, ... | | Authors: | Wu, Z, Liu, D, Shen, H, Ji, Q. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure-directed functional evolution of the miniature CRISPR-AsCas12f1 system
To Be Published
|
|
2V55
 
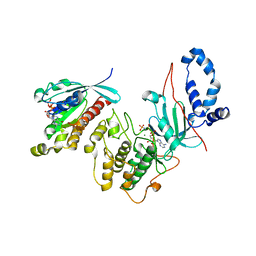 | | Mechanism of multi-site phosphorylation from a ROCK-I:RhoE complex structure | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Komander, D, Garg, R, Wan, P.T.C, Ridley, A.J, Barford, D. | | Deposit date: | 2008-10-01 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.705 Å) | | Cite: | Mechanism of Multi-Site Phosphorylation from a Rock-I:Rhoe Complex Structure.
Embo J., 27, 2008
|
|
6PPH
 
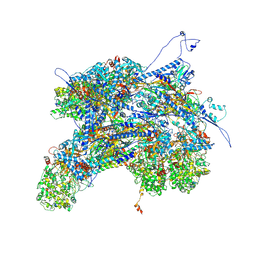 | | Kaposi's sarcoma-associated herpesvirus (KSHV), C1 penton vertex register, CATC-binding structure | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Gong, D, Dai, X, Jih, J, Liu, Y.T, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2019-07-06 | | Release date: | 2019-09-11 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | DNA-Packing Portal and Capsid-Associated Tegument Complexes in the Tumor Herpesvirus KSHV.
Cell, 178, 2019
|
|
1CF4
 
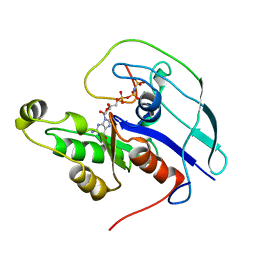 | | CDC42/ACK GTPASE-BINDING DOMAIN COMPLEX | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, PROTEIN (ACTIVATED P21CDC42HS KINASE), ... | | Authors: | Mott, H.R, Owen, D, Nietlispach, D, Lowe, P.N, Lim, L, Laue, E.D. | | Deposit date: | 1999-03-23 | | Release date: | 1999-06-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the small G protein Cdc42 bound to the GTPase-binding domain of ACK.
Nature, 399, 1999
|
|
1BWE
 
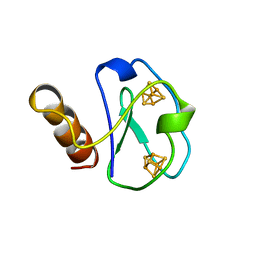 | | ARTIFICIAL FE8S8 FERREDOXIN: THE D13C VARIANT OF BACILLUS SCHLEGELII FE7S8 FERREDOXIN | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Aono, S, Bentrop, D, Bertini, I, Cosenza, G, Luchinat, C. | | Deposit date: | 1998-09-23 | | Release date: | 1998-09-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an artificial Fe8S8 ferredoxin: the D13C variant of Bacillus schlegelii Fe7S8 ferredoxin.
Eur.J.Biochem., 258, 1998
|
|
1S53
 
 | | Thr46Ser Bacteriorhodopsin | | Descriptor: | RETINAL, bacteriorhodopsin | | Authors: | Yohannan, S, Faham, S, Yang, D, Grosfeld, D, Chamberlain, A.K, Bowie, J.U. | | Deposit date: | 2004-01-19 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A C(alpha)-H.O Hydrogen Bond in a Membrane Protein Is Not Stabilizing
J.Am.Chem.Soc., 126, 2004
|
|
7W5L
 
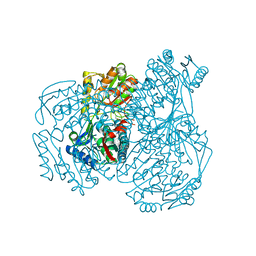 | | The crystal structure of the oxidized form of Gluconobacter oxydans WSH-004 SNDH | | Descriptor: | L-sorbosone dehydrogenase, NAD(P) dependent | | Authors: | Li, D, Hou, X.D, Rao, Y.J, Zhou, J.W, Chen, J. | | Deposit date: | 2021-11-30 | | Release date: | 2023-01-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insight into the Catalytic Mechanisms of an L-Sorbosone Dehydrogenase.
Adv Sci, 10, 2023
|
|
7WQV
 
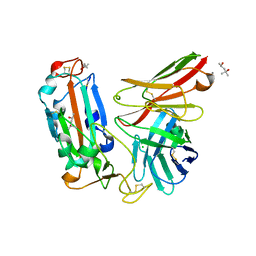 | | Crystal structure of a neutralizing monoclonal antibody (Ab08) in complex with SARS-CoV-2 receptor-binding domain (RBD) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ab08, ... | | Authors: | Zha, J, Meng, L, Zhang, X, Li, D. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Spike-destructing human antibody effectively neutralizes Omicron-included SARS-CoV-2 variants with therapeutic efficacy.
Plos Pathog., 19, 2023
|
|
6PPI
 
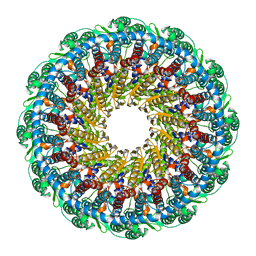 | | Kaposi's sarcoma-associated herpesvirus (KSHV), C12 portal dodecamer structure | | Descriptor: | Portal protein | | Authors: | Gong, D, Dai, X, Jih, J, Liu, Y.T, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2019-07-07 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | DNA-Packing Portal and Capsid-Associated Tegument Complexes in the Tumor Herpesvirus KSHV.
Cell, 178, 2019
|
|
5MU2
 
 | | ACC1 Fab fragment in complex with CII583-591 (CG10) | | Descriptor: | ACC1 Fab fragment heavy chain, ACC1 Fab fragment light chain, synthetic peptide containing the CII583-591 epitope of collagen type II | | Authors: | Dobritzsch, D, Holmdahl, R, Ge, C. | | Deposit date: | 2017-01-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Anti-citrullinated protein antibodies cause arthritis by cross-reactivity to joint cartilage.
JCI Insight, 2, 2017
|
|
1C9Y
 
 | | HUMAN ORNITHINE TRANSCARBAMYLASE: CRYSTALLOGRAPHIC INSIGHTS INTO SUBSTRATE RECOGNITION AND CATALYTIC MECHANISM | | Descriptor: | NORVALINE, ORNITHINE CARBAMOYLTRANSFERASE, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Shi, D, Yu, X, Morizono, H, Tuchman, M, Allewell, N.M. | | Deposit date: | 1999-08-03 | | Release date: | 2000-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human ornithine transcarbamylase complexed with carbamoyl phosphate and L-norvaline at 1.9 A resolution.
Proteins, 39, 2000
|
|
3ZO8
 
 | | Wild-type chorismate mutase of Bacillus subtilis at 1.6 A resolution | | Descriptor: | CHORISMATE MUTASE AROH | | Authors: | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | Deposit date: | 2013-02-20 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7W5N
 
 | | The crystal structure of the reduced form of Gluconobacter oxydans WSH-004 SNDH | | Descriptor: | L-sorbosone dehydrogenase, NAD(P) dependent, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, D, Hou, X.D, Rao, Y.J, Yin, D.J, Zhou, J.W, Chen, J. | | Deposit date: | 2021-11-30 | | Release date: | 2023-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.988 Å) | | Cite: | Structural Insight into the Catalytic Mechanisms of an L-Sorbosone Dehydrogenase.
Adv Sci, 10, 2023
|
|
6PPD
 
 | | Kaposi's sarcoma-associated herpesvirus (KSHV), C1 penton vertex register, CATC-absent structure | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Gong, D, Dai, X, Jih, J, Liu, Y.T, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2019-07-06 | | Release date: | 2019-09-11 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | DNA-Packing Portal and Capsid-Associated Tegument Complexes in the Tumor Herpesvirus KSHV.
Cell, 178, 2019
|
|
1A0K
 
 | |
1RNW
 
 | | RECOMBINANT RIBONUCLEASE A CRYSTALLIZED FROM 80% AMMONIUM SULPHATE | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Fedorov, A.A, Joseph-Mccarthy, D, Fedorov, E.V, Sirakova, D, Graf, I, Almo, S.C. | | Deposit date: | 1996-11-08 | | Release date: | 1997-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ionic interactions in crystalline bovine pancreatic ribonuclease A.
Biochemistry, 35, 1996
|
|
7USA
 
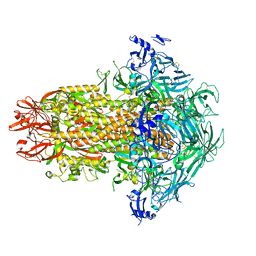 | | Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the swung out conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
3ZJF
 
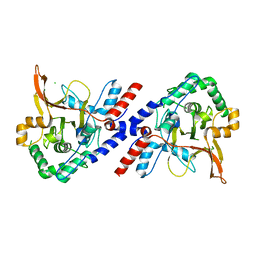 | | A20 OTU domain with irreversibly oxidised Cys103 from 270 min H2O2 soak. | | Descriptor: | A20P50, CHLORIDE ION | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
5N21
 
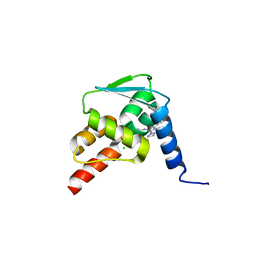 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | Descriptor: | 2-[(2~{S})-1-[3-cyano-7-[(2-oxidanylidene-3,4-dihydro-1~{H}-quinolin-6-yl)amino]pyrazolo[1,5-a]pyrimidin-5-yl]pyrrolidin-2-yl]ethanoic acid, B-cell lymphoma 6 protein, CHLORIDE ION | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
1R17
 
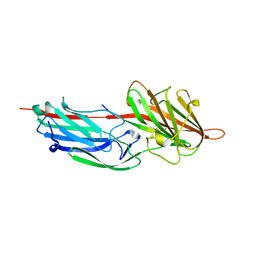 | | Crystal Structure Analysis of S.epidermidis adhesin SdrG binding to Fibrinogen (adhesin-ligand complex) | | Descriptor: | CALCIUM ION, fibrinogen-binding protein SdrG, fibrinopeptide B | | Authors: | Ponnuraj, K, Bowden, M.G, Davis, S, Gurusiddappa, S, Moore, D, Choe, D, Xu, Y, Hook, M, Narayana, S.V.L. | | Deposit date: | 2003-09-23 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A "dock, lock and latch" Structural Model for a Staphylococcal Adhesin Binding to Fibrinogen
Cell(Cambridge,Mass.), 115, 2003
|
|
3ZNX
 
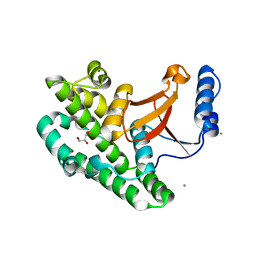 | | Crystal structure of the OTU domain of OTULIN D336A mutant | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Keusekotten, K, Elliott, P.R, Glockner, L, Kulathu, Y, Wauer, T, Krappmann, D, Hofmann, K, Komander, D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Otulin Antagonizes Lubac Signaling by Specifically Hydrolyzing met1-Linked Polyubiquitin.
Cell(Cambridge,Mass.), 153, 2013
|
|
1R4L
 
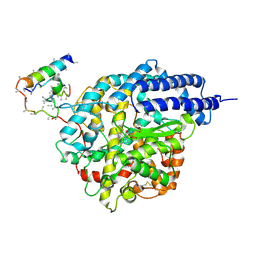 | | Inhibitor Bound Human Angiotensin Converting Enzyme-Related Carboxypeptidase (ACE2) | | Descriptor: | (S,S)-2-{1-CARBOXY-2-[3-(3,5-DICHLORO-BENZYL)-3H-IMIDAZOL-4-YL]-ETHYLAMINO}-4-METHYL-PENTANOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Towler, P, Staker, B, Prasad, S.G, Menon, S, Ryan, D, Tang, J, Parsons, T, Fisher, M, Williams, D, Dales, N.A, Patane, M.A, Pantoliano, M.W. | | Deposit date: | 2003-10-07 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis.
J.Biol.Chem., 279, 2004
|
|
7UCP
 
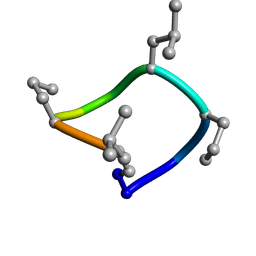 | | computationally designed macrocycle | | Descriptor: | computationally designed cyclic peptide D8.3.p2 | | Authors: | Bhardwaj, G, Baker, D, Rettie, S, Glynn, C, Sawaya, M. | | Deposit date: | 2022-03-17 | | Release date: | 2022-09-14 | | Last modified: | 2022-09-28 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Accurate de novo design of membrane-traversing macrocycles.
Cell, 185, 2022
|
|
1R7G
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 100mM DPC) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R61
 
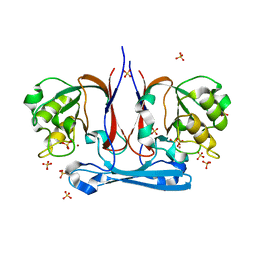 | | The structure of predicted metal-dependent hydrolase from Bacillus stearothermophilus | | Descriptor: | SULFATE ION, ZINC ION, metal-dependent hydrolase | | Authors: | Maderova, J, Borek, D, Tomchick, D, Joachimiak, A, Collart, F, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-14 | | Release date: | 2004-03-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of potential metal-dependent hydrolase with cyclase activity
To be Published
|
|
