3F6C
 
 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI | | Descriptor: | GLYCEROL, Positive transcription regulator evgA | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Wu, B, Bain, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI
To be Published
|
|
2KSJ
 
 | | Structure and Dynamics of the Membrane-bound form of Pf1 Coat Protein: Implications for Structural Rearrangement During Virus Assembly | | Descriptor: | Capsid protein G8P | | Authors: | Park, S, Marassi, F, Black, D, Opella, S.J. | | Deposit date: | 2010-01-05 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Structure and dynamics of the membrane-bound form of Pf1 coat protein: implications of structural rearrangement for virus assembly.
Biophys.J., 99, 2010
|
|
2KUM
 
 | | Solution structure of the human chemokine CCL27 | | Descriptor: | C-C motif chemokine 27 | | Authors: | Kirkpatrick, J.P, Jansma, A, Hsu, A, Handel, T.M, Nietlispach, D. | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR analysis of the structure, dynamics, and unique oligomerization properties of the chemokine CCL27.
J.Biol.Chem., 285, 2010
|
|
4AC4
 
 | | CRYSTAL STRUCTURE OF TRANSTHYRETIN IN COMPLEX WITH LIGAND C-18 | | Descriptor: | 3-METHOXY-4-PHENOXYBENZOIC ACID, TRANSTHYRETIN | | Authors: | Tomar, D, Khan, T, Singh, R.R, Mishra, S, Gupta, S, Surolia, A, Salunke, D.M. | | Deposit date: | 2011-12-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Study of Novel Transthyretin Ligands Exhibiting Negative-Cooperativity between Two Thyroxine Binding Sites.
Plos One, 7, 2012
|
|
2KVZ
 
 | | Structure of residues 161-235 of putative peptidoglycan binding protein lmo0835 from Listeria monocytogenes: target LmR64B of the Northeast Structural Genomics Consortium | | Descriptor: | IspE | | Authors: | Cort, J.R, Ramelot, T.A, Lee, D, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-29 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of residues 161-235 of putative peptidoglycan binding protein lmo0835 from Listeria monocytogenes: target LmR64B of the Northeast Structural Genomics Consortium
To be Published
|
|
3SBB
 
 | | Disulphide-mediated Tetramer of T4 Lysozyme R76C/R80C by Synthetic Symmetrization | | Descriptor: | CHLORIDE ION, Lysozyme | | Authors: | Laganowsky, A, Soriaga, A.B, Zhao, M, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-03 | | Release date: | 2011-09-21 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (1.434 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
4AM4
 
 | | Bacterioferritin from Blastochloris viridis | | Descriptor: | BACTERIOFERRITIN, FE (III) ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wahlgren, W.Y, Omran, H, von Stetten, D, Royant, A, van der Post, S, Katona, G. | | Deposit date: | 2012-03-07 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Characterization of Bacterioferritin from Blastochloris Viridis.
Plos One, 7, 2012
|
|
3EWU
 
 | | D312N mutant of human orotidyl-5'-monophosphate decarboxylase in complex with 6-acetyl-UMP, covalent adduct | | Descriptor: | 6-ethyluridine 5'-phosphate, GLYCEROL, Orotidine-5'-phosphate decarboxylase, ... | | Authors: | Heinrich, D, Wittmann, J, Diederichsen, U, Rudolph, M. | | Deposit date: | 2008-10-16 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Lys314 is a nucleophile in non-classical reactions of orotidine-5'-monophosphate decarboxylase
Chemistry, 15, 2009
|
|
3F3V
 
 | | Kinase domain of cSrc in complex with inhibitor RL45 (Type II) | | Descriptor: | 1-{4-[(6-aminoquinazolin-4-yl)amino]phenyl}-3-[3-tert-butyl-1-(3-methylphenyl)-1H-pyrazol-5-yl]urea, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Grutter, C, Kluter, S, Getlik, M, Rauh, D. | | Deposit date: | 2008-10-31 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hybrid compound design to overcome the gatekeeper T338M mutation in cSrc
J.Med.Chem., 52, 2009
|
|
2KXH
 
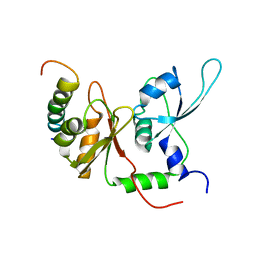 | | Solution structure of the first two RRM domains of FIR in the complex with FBP Nbox peptide | | Descriptor: | Poly(U)-binding-splicing factor PUF60, peptide of Far upstream element-binding protein 1 | | Authors: | Cukier, C.D, Ramos, A, Hollingworth, D, Diaz-Moreno, I, Kelly, G. | | Deposit date: | 2010-05-05 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of FIR-mediated c-myc transcriptional control.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3F6E
 
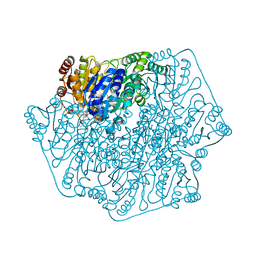 | | Crystal structure of benzoylformate decarboxylase in complex with the pyridyl inhibitor 3-PKB | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-2-[(1S,2E)-1-hydroxy-3-pyridin-3-ylprop-2-en-1-yl]-4-methyl-1,3-thiazol-3-ium, Benzoylformate decarboxylase, MAGNESIUM ION | | Authors: | Brandt, G.S, McLeish, M.J, Kenyon, G.L, Petsko, G.A, Ringe, D, Jordan, F. | | Deposit date: | 2008-11-05 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Detection and time course of formation of major thiamin diphosphate-bound covalent intermediates derived from a chromophoric substrate analogue on benzoylformate decarboxylase.
Biochemistry, 48, 2009
|
|
3W79
 
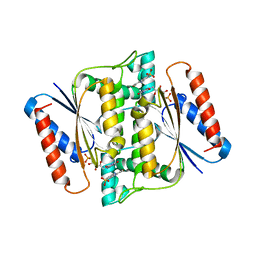 | | Crystal Structure of azoreductase AzrC in complex with sulfone-modified azo dye Orange I | | Descriptor: | 4-[(E)-(4-hydroxynaphthalen-1-yl)diazenyl]benzenesulfonic acid, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | Authors: | Ogata, D, Yu, J, Ooi, T, Yao, M. | | Deposit date: | 2013-02-27 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of AzrA and of AzrC complexed with substrate or inhibitor: insight into substrate specificity and catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5VRI
 
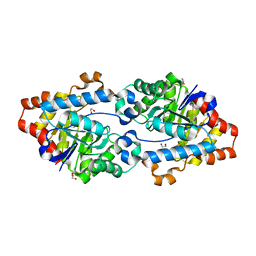 | | Crystal structure of SsoPox AsA6 mutant (F46L-C258A-W263M-I280T) - closed form | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Hiblot, J, Gotthard, G, Jacquet, P, Daude, D, Bergonzi, C, Chabriere, E, Elias, M. | | Deposit date: | 2017-05-10 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Rational engineering of a native hyperthermostable lactonase into a broad spectrum phosphotriesterase.
Sci Rep, 7, 2017
|
|
3SI0
 
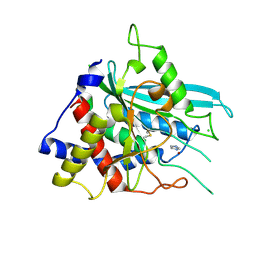 | | Structure of glycosylated human glutaminyl cyclase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glutaminyl-peptide cyclotransferase, ... | | Authors: | Parthier, C, Carrillo, D, Stubbs, M.T. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Glycosylated Mammalian Glutaminyl Cyclases Reveal Conformational Variability near the Active Center.
Biochemistry, 50, 2011
|
|
2KW7
 
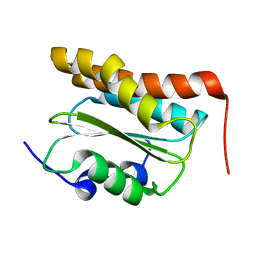 | | Solution NMR Structure of the N-terminal domain of protein PG_0361 from P.gingivalis, Northeast Structural Genomics Consortium Target PgR37A | | Descriptor: | Conserved domain protein | | Authors: | Eletsky, A, Mills, J.L, Lee, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the N-terminal domain of protein PG_0361 from P.gingivalis, Northeast Structural Genomics Consortium Target Target PgR37A
To be Published
|
|
2KXL
 
 | |
5VYH
 
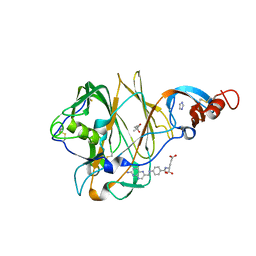 | | Crystal Structure of MERS-CoV S1 N-terminal Domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Wang, N, Wrapp, D, Pallesen, J, Ward, A.B, McLellan, J.S. | | Deposit date: | 2017-05-25 | | Release date: | 2017-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VZ5
 
 | | Crystal structure of an anaplastic lymphoma kinase-derived neuroblastoma tumor antigen bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Toor, J, Rao, A.A, Salama, S, Tripathi, S, Haussler, D, Sgourakis, N.G. | | Deposit date: | 2017-05-26 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5901 Å) | | Cite: | A Recurrent Mutation in Anaplastic Lymphoma Kinase with Distinct Neoepitope Conformations.
Front Immunol, 9, 2018
|
|
2KT7
 
 | | Solution NMR structure of mucin-binding domain of protein lmo0835 from Listeria monocytogenes, Northeast Structural Genomics Consortium Target LmR64A | | Descriptor: | Putative peptidoglycan bound protein (LPXTG motif) | | Authors: | Eletsky, A, He, Y, Lee, D, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-20 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of mucin-binding domain of protein lmo0835 from Listeria monocytogenes
To be Published
|
|
2KRT
 
 | | Solution NMR Structure of a Conserved Hypothetical Membrane Lipoprotein Obtained from Ureaplasma parvum: Northeast Structural Genomics Consortium Target UuR17A (139-239) | | Descriptor: | Conserved hypothetical membrane lipoprotein | | Authors: | Mani, R, Swapna, G, Janjua, H, Ciccosanti, C, Huang, Y, Patel, D, Xiao, R, Acton, T, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-22 | | Release date: | 2010-01-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a Conserved Hypothetical Membrane Lipoprotein obtained from Ureaplasma parvum : Northeast Structural Genomics Consortium Target UuR17A (139-239)
To be Published
|
|
3EZ3
 
 | | Crystal Structure of Plasmodium vivax geranylgeranylpyrophosphate synthase PVX_092040 with zoledronate and IPP bound | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl pyrophosphate synthase, ... | | Authors: | Wernimont, A.K, Lew, J, Zhao, Y, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-22 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Crystal Structure of Plasmodium vivax geranylgeranylpyrophosphate synthase PVX_092040 with zoledronate and IPP bound
TO BE PUBLISHED
|
|
3WDE
 
 | |
3WFR
 
 | | tRNA processing enzyme complex 2 | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
2KS6
 
 | |
2KSY
 
 | | Solution nmr structure of sensory rhodopsin II | | Descriptor: | RETINAL, Sensory rhodopsin II | | Authors: | Gautier, A, Mott, H.R, Bostock, M.J, Kirkpatrick, J.P, Nietlispach, D. | | Deposit date: | 2010-01-14 | | Release date: | 2010-06-02 | | Last modified: | 2015-06-10 | | Method: | SOLUTION NMR | | Cite: | Structure determination of the seven-helix transmembrane receptor sensory rhodopsin II by solution NMR spectroscopy.
Nat.Struct.Mol.Biol., 17, 2010
|
|
