1JCP
 
 | | Solution structure of the lactam analogue EDap of HIV gp41 600-612 loop. | | Descriptor: | Edap : ACE-Ile-Trp-Glu-Ser-Gly-Lys-Leu-Ile-Dap-Thr-Thr-Ala ANALOGUE OF HIV GP41 | | Authors: | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | Deposit date: | 2001-06-11 | | Release date: | 2003-07-01 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
5CR1
 
 | | Crystal structure of TTR/resveratrol/T4 complex | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, RESVERATROL, Transthyretin | | Authors: | Zanotti, G, Florio, P, Folli, C, Cianci, M, Del Rio, D, Berni, R. | | Deposit date: | 2015-07-22 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.545 Å) | | Cite: | Transthyretin Binding Heterogeneity and Anti-amyloidogenic Activity of Natural Polyphenols and Their Metabolites.
J.Biol.Chem., 290, 2015
|
|
7TFP
 
 | | Human Ornithine Aminotransferase cocrystallized with its inhibitor, (1S,3S)-3-amino-4-(difluoromethylene)cyclopentane-1-carboxylic acid. | | Descriptor: | (1S,3S,4S)-3-amino-4-(fluoromethyl)cyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Butrin, A, Zhu, W, Silverman, R, Liu, D. | | Deposit date: | 2022-01-06 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Rational Design, Synthesis, and Mechanism of (3 S ,4 R )-3-Amino-4-(difluoromethyl)cyclopent-1-ene-1-carboxylic Acid: Employing a Second-Deprotonation Strategy for Selectivity of Human Ornithine Aminotransferase over GABA Aminotransferase.
J.Am.Chem.Soc., 144, 2022
|
|
5CQ1
 
 | | Disproportionating enzyme 1 from Arabidopsis - cycloamylose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
5CSB
 
 | | The crystal structure of beta2-microglobulin D76N mutant at room temperature | | Descriptor: | Beta-2-microglobulin | | Authors: | de Rosa, M, Mota, C.S, de Sanctis, D, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity.
Nat Commun, 9, 2018
|
|
1JCO
 
 | | Solution structure of the monomeric [Thr(B27)->Pro,Pro(B28)->Thr] insulin mutant (PT insulin) | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Keller, D, Clausen, R, Josefsen, K, Led, J.J. | | Deposit date: | 2001-06-11 | | Release date: | 2001-10-03 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Flexibility and bioactivity of insulin: an NMR investigation of the solution structure and folding of an unusually flexible human insulin mutant with increased biological activity.
Biochemistry, 40, 2001
|
|
5DCN
 
 | | Crystal structure of LC3 in complex with TECPR2 LIR | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Stadel, D, Huber, J, Dotsch, V, Rogov, V.V, Behrends, C, Akutsu, M. | | Deposit date: | 2015-08-24 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TECPR2 Cooperates with LC3C to Regulate COPII-Dependent ER Export.
Mol.Cell, 60, 2015
|
|
5CW9
 
 | | Crystal structure of De novo designed ferredoxin-ferredoxin domain insertion protein | | Descriptor: | De novo designed ferredoxin-ferredoxin domain insertion protein | | Authors: | DiMaio, F, King, I.C, Gleixner, J, Doyle, L, Stoddard, B, Baker, D. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.108 Å) | | Cite: | Precise assembly of complex beta sheet topologies from de novo designed building blocks.
Elife, 4, 2015
|
|
1JAK
 
 | | Streptomyces plicatus beta-N-acetylhexosaminidase in Complex with (2R,3R,4S,5R)-2-acetamido-3,4-dihydroxy-5-hydroxymethyl-piperidinium chloride (IFG) | | Descriptor: | (2R,3R,4S,5R)-2-ACETAMIDO-3,4-DIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Mark, B.L, Vocadlo, D.J, Zhao, D, Knapp, S, Withers, S.G, James, M.N. | | Deposit date: | 2001-05-30 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical and structural assessment of the 1-N-azasugar GalNAc-isofagomine as a potent family 20 beta-N-acetylhexosaminidase inhibitor.
J.Biol.Chem., 276, 2001
|
|
1LVH
 
 | | The Structure of Phosphorylated beta-phosphoglucomutase from Lactoccocus lactis to 2.3 angstrom resolution | | Descriptor: | MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2002-05-28 | | Release date: | 2002-08-14 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Caught in the act: the structure of phosphorylated beta-phosphoglucomutase from Lactococcus lactis.
Biochemistry, 41, 2002
|
|
5D93
 
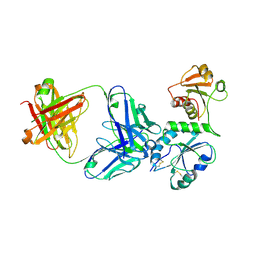 | |
1JN2
 
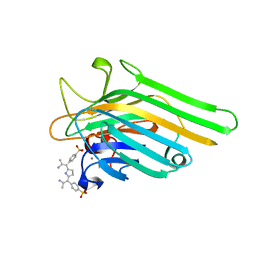 | | Crystal Structure of meso-tetrasulphonatophenyl porphyrin complexed with Concanavalin A | | Descriptor: | 5,10,15,20-TETRAKIS(4-SULPFONATOPHENYL)-21H,23H-PORPHINE, CALCIUM ION, Concanavalin-A, ... | | Authors: | Goel, M, Jain, D, Kaur, K.J, Kenoth, R, Maiya, B.G, Swamy, M.J, Salunke, D.M. | | Deposit date: | 2001-07-22 | | Release date: | 2003-07-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional equality in the absence of structural similarity: an added dimension to
molecular mimicry
J.Biol.Chem., 276, 2000
|
|
1JR6
 
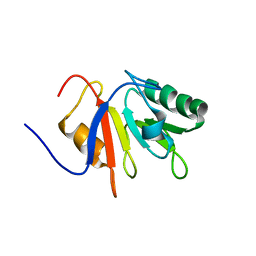 | |
1NOG
 
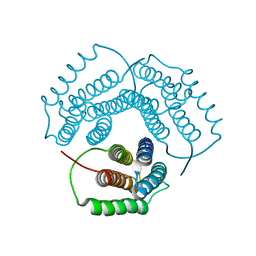 | | Crystal Structure of Conserved Protein 0546 from Thermoplasma Acidophilum | | Descriptor: | conserved hypothetical protein TA0546 | | Authors: | Saridakis, V, Sanishvili, R, Iakounine, A, Xu, X, Pennycooke, M, Gu, J, Joachimiak, A, Arrowsmith, C.H, Edwards, A.M, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-16 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structural basis for methylmalonic aciduria. The crystal structure of archaeal ATP:cobalamin adenosyltransferase.
J.Biol.Chem., 279, 2004
|
|
5DBK
 
 | |
1JTQ
 
 | | E. coli TS Complex with dUMP and the Pyrrolo(2,3-d)pyrimidine-based Antifolate LY341770 | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-4-(2H-TETRAZOL-5-YL)-BUTYRIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Sayre, P.H, Finer-Moore, J.S, Fritz, T.A, Biermann, D, Gates, S.B, MacKellar, W.C, Patel, V.F, Stroud, R.M. | | Deposit date: | 2001-08-21 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multi-targeted antifolates aimed at avoiding drug resistance form covalent closed inhibitory complexes with human and Escherichia coli thymidylate synthases.
J.Mol.Biol., 313, 2001
|
|
5DEJ
 
 | | Transthyretin natural mutant A19D | | Descriptor: | CALCIUM ION, Transthyretin | | Authors: | Pereira, H.M, Ferreira, P, Andrade, C, Lima, C, Palhano, F, Foguel, D. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of human Transthyretin natural mutant A19D
To Be Published
|
|
1JYI
 
 | | CONCANAVALIN A/12-MER PEPTIDE COMPLEX | | Descriptor: | 12-mer peptide, CALCIUM ION, Concanavalin-Br, ... | | Authors: | Jain, D, Kaur, K.J, Sundaravadivel, B, Salunke, D.M. | | Deposit date: | 2001-09-12 | | Release date: | 2002-09-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and Functional Consequences of Peptide-carbohydrate Mimicry. Crystal Structure of a Carbohydrate-mimicking Peptide Bound to Concanavalin A.
J.Biol.Chem., 275, 2000
|
|
1JR3
 
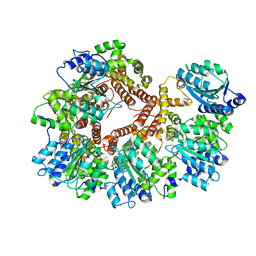 | | Crystal Structure of the Processivity Clamp Loader Gamma Complex of E. coli DNA Polymerase III | | Descriptor: | DNA polymerase III subunit gamma, DNA polymerase III, delta subunit, ... | | Authors: | Jeruzalmi, D, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2001-08-10 | | Release date: | 2001-09-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 106, 2001
|
|
1K0I
 
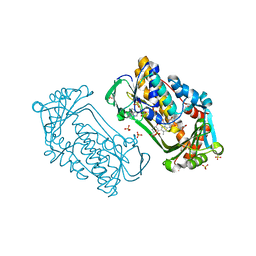 | | Pseudomonas aeruginosa phbh R220Q in complex with 100mM PHB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID, ... | | Authors: | Wang, J, Ortiz-Maldonado, M, Entsch, B, Ballou, D, Gatti, D.L. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein and ligand dynamics in 4-hydroxybenzoate hydroxylase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JTS
 
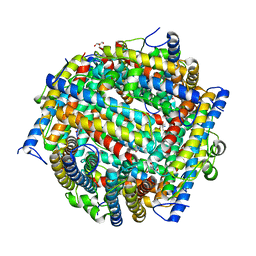 | |
5D6F
 
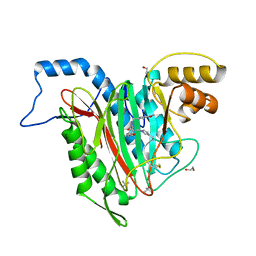 | | Structure of human methionine aminopeptidase-2 complexed with spiroepoxytriazole inhibitor (+)-31b | | Descriptor: | (4S,7R)-7-hydroxy-1-(4-methoxybenzyl)-7-methyl-4,5,6,7-tetrahydro-1H-benzotriazol-4-yl propan-2-ylcarbamate, 1,2-ETHANEDIOL, COBALT (II) ION, ... | | Authors: | Janowski, R, Miller, A.K, Niessing, D. | | Deposit date: | 2015-08-12 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Spiroepoxytriazoles Are Fumagillin-like Irreversible Inhibitors of MetAP2 with Potent Cellular Activity.
Acs Chem.Biol., 11, 2016
|
|
1JPE
 
 | |
1JNI
 
 | | Structure of the NapB subunit of the periplasmic nitrate reductase from Haemophilus influenzae. | | Descriptor: | DIHEME CYTOCHROME C NAPB, HEME C | | Authors: | Brige, A, Leys, D, Meyer, T.E, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2001-07-24 | | Release date: | 2002-05-17 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The 1.25 A resolution structure of the diheme NapB subunit of soluble nitrate reductase reveals a novel cytochrome c fold with a stacked heme arrangement.
Biochemistry, 41, 2002
|
|
5EA4
 
 | | Crystal Structure of Inhibitor JNJ-49153390 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 3-[[5-bromanyl-1-(3-methylsulfonylpropyl)benzimidazol-2-yl]methyl]-1-cyclopropyl-imidazo[4,5-c]pyridin-2-one, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
