6TX5
 
 | |
3T4N
 
 | | Structure of the regulatory fragment of Saccharomyces cerevisiae AMPK in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmdit, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-07-26 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
6TXX
 
 | | CRYSTAL STRUCTURE OF HUMAN FKBP51 FK1 DOMAIN A19T MUTANT IN COMPLEX WITH SAFit2 | | Descriptor: | (1R)-3-(3,4-dimethoxyphenyl)-1-{3-[2-(morpholin-4-yl)ethoxy]phenyl}propyl (2S)-1-[(2S)-2-[(1S)-cyclohex-2-en-1-yl]-2-(3,4,5-trimethoxyphenyl)acetyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Fiegen, D, Draxler, S.W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Hybrid Screening Approach for Very Small Fragments: X-ray and Computational Screening on FKBP51.
J.Med.Chem., 63, 2020
|
|
5EIV
 
 | | Crystal structure of complex of osteoclast-associated immunoglobulin-like receptor (OSCAR) and a synthetic collagen consensus peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | Authors: | Zhou, L, Blaszczyk, M, Chirgadze, D, Bihan, D, Farndale, R.W. | | Deposit date: | 2015-10-30 | | Release date: | 2015-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | Structural basis for collagen recognition by the immune receptor OSCAR.
Blood, 127, 2016
|
|
3SPA
 
 | | Crystal Structure of Human Mitochondrial RNA Polymerase | | Descriptor: | CHLORIDE ION, DNA-directed RNA polymerase, mitochondrial, ... | | Authors: | Ringel, R, Sologub, M, Morozov, Y.I, Litonin, D, Cramer, P, Temiakov, D. | | Deposit date: | 2011-07-01 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of human mitochondrial RNA polymerase
Nature, 478, 2011
|
|
6UDT
 
 | | X-ray co-crystal structure of compound 10 bound to human Mcl-1 | | Descriptor: | (4S,7aR,9aR,10S,11E,18R)-6'-chloro-10,18-dihydroxy-15-methyl-16-oxo-3',4',7,7a,8,9,9a,10,13,14,15,16,17,18-tetradecahydro-2'H,3H,5H-spiro[1,19-(ethanediylidene)cyclobuta[n][1,4]oxazepino[4,3-a][1,8]diazacyclohexadecine-4,1'-naphthalene]-18-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X, Whittington, D. | | Deposit date: | 2019-09-19 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery and in Vivo Evaluation of Macrocyclic Mcl-1 Inhibitors Featuring an alpha-Hydroxy Phenylacetic Acid Pharmacophore or Bioisostere.
J.Med.Chem., 62, 2019
|
|
6UDU
 
 | | X-ray co-crystal structure of compound 8 bound to human Mcl-1 | | Descriptor: | (4S,11E,17R)-6'-chloro-17-hydroxy-14-methyl-15-oxo-3',4',8,9,10,13,14,15,16,17-decahydro-2'H,3H,5H,7H-spiro[1,18-(ethanediylidene)[1,4]oxazepino[4,3-a][1,8]diazacyclopentadecine-4,1'-naphthalene]-17-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X, Whittington, D. | | Deposit date: | 2019-09-19 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery and in Vivo Evaluation of Macrocyclic Mcl-1 Inhibitors Featuring an alpha-Hydroxy Phenylacetic Acid Pharmacophore or Bioisostere.
J.Med.Chem., 62, 2019
|
|
6UDV
 
 | | X-ray co-crystal structure of compound 3 bound to human Mcl-1 | | Descriptor: | (4S,7aR,9aR,10S,11E,14S,15R)-6'-chloro-10-hydroxy-14,15-dimethyl-3',4',7a,8,9,9a,10,13,14,15-decahydro-2'H,3H,5H-spiro[1,19-(ethanediylidene)-16lambda~6~-cyclobuta[i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-4,1'-naphthalene]-16,16,18(7H,17H)-trione, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X, Whittington, D. | | Deposit date: | 2019-09-19 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery and in Vivo Evaluation of Macrocyclic Mcl-1 Inhibitors Featuring an alpha-Hydroxy Phenylacetic Acid Pharmacophore or Bioisostere.
J.Med.Chem., 62, 2019
|
|
4MH7
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC1896 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-butyl-2-(butylamino)-4-[(trans-4-hydroxycyclohexyl)amino]-N-methylpyrimidine-5-carboxamide, ... | | Authors: | Zhang, W, McIver, A, Stashko, M.A, Deryckere, D, Branchford, B.R, Hunter, D, Kireev, D.B, Miley, M.J, Norris-Drouin, J, Stewart, W.M, Lee, M, Sather, S, Zhou, Y, DiPaola, J.A, Machius, M, Janzen, W.P, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2013-08-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Mer specific tyrosine kinase inhibitors for the treatment and prevention of thrombosis.
J.Med.Chem., 56, 2013
|
|
2QZV
 
 | | Draft Crystal Structure of the Vault Shell at 9 Angstroms Resolution | | Descriptor: | Major vault protein | | Authors: | Anderson, D.H, Kickhoefer, V.A, Sievers, S.A, Rome, L.H, Eisenberg, D. | | Deposit date: | 2007-08-17 | | Release date: | 2007-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (9 Å) | | Cite: | Draft crystal structure of the vault shell at 9-A resolution.
Plos Biol., 5, 2007
|
|
4DN8
 
 | | Structure of porcine surfactant protein D neck and carbohydrate recognition domain complexed with mannose | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D, beta-D-mannopyranose | | Authors: | van Eijk, M, Rynkiewicz, M.J, White, M.R, Hartshorn, K.L, Zou, X, Schulten, K, Luo, D, Crouch, E.C, Cafarella, T.M, Head, J.F, Haagsman, H.P, Seaton, B.A. | | Deposit date: | 2012-02-08 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Unique Sugar-binding Site Mediates the Distinct Anti-influenza Activity of Pig Surfactant Protein D.
J.Biol.Chem., 287, 2012
|
|
2FBW
 
 | | Avian respiratory complex II with carboxin bound | | Descriptor: | (~{Z})-2-oxidanylbut-2-enedioic acid, 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, AZIDE ION, ... | | Authors: | Huang, L.S, Sun, G, Cobessi, D, Wang, A.C, Shen, J.T, Tung, E.Y, Anderson, V.E, Berry, E.A. | | Deposit date: | 2005-12-10 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme.
J.Biol.Chem., 281, 2006
|
|
4E0O
 
 | | SVQIVYK segment from human Tau (305-311) displayed on 54-membered macrocycle scaffold (form III) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide SVQIVYK(ORN)EF(HAO)(4BF)K(ORN), PHOSPHATE ION | | Authors: | Zhao, M, Liu, C, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2012-03-04 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Out-of-register beta-sheets suggest a pathway to toxic amyloid aggregates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7XJ2
 
 | | Structure of human TRPV3_G573S in complex with Trpvicin in C4 symmetry | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ1
 
 | | Structure of human TRPV3_G573S in complex with Trpvicin in C2 symmetry | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ3
 
 | | Structure of human TRPV3 | | Descriptor: | [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate, fusion of transient receptor potential cation channel subfamily V member 3 and 3C-GFP | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ0
 
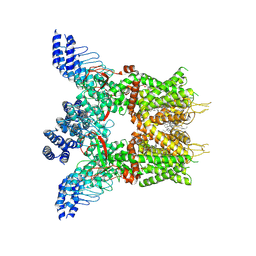 | | Structure of human TRPV3 in complex with Trpvicin | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7P36
 
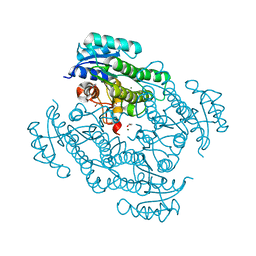 | | X-ray structure of Lactobacillus kefir alcohol dehydrogenase (wild type) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bischoff, D, Walla, B, Janowski, R, Niessing, D, Weuster-Botz, D. | | Deposit date: | 2021-07-07 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Transfer of a Rational Crystal Contact Engineering Strategy between Diverse Alcohol Dehydrogenases
Crystals, 11, 2021
|
|
7XNK
 
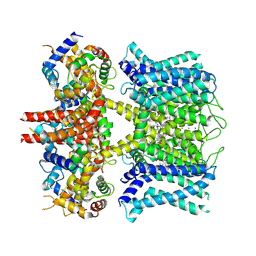 | | human KCNQ1-CaM in complex with ML277 | | Descriptor: | (2R)-N-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-1-(4-methylbenzene-1-sulfonyl)piperidine-2-carboxamide, Calmodulin-3, POTASSIUM ION, ... | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mechanisms for the activation of human cardiac KCNQ1 channel by electro-mechanical coupling enhancers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XNI
 
 | | human KCNQ1-CaM in apo state | | Descriptor: | Calmodulin-3, Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanisms for the activation of human cardiac KCNQ1 channel by electro-mechanical coupling enhancers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XNL
 
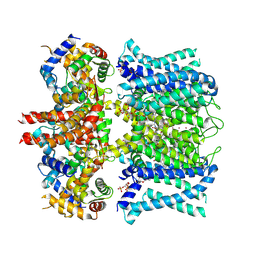 | | human KCNQ1-CaM-ML277-PIP2 complex in state A | | Descriptor: | (2R)-N-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-1-(4-methylbenzene-1-sulfonyl)piperidine-2-carboxamide, Calmodulin-3, POTASSIUM ION, ... | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural mechanisms for the activation of human cardiac KCNQ1 channel by electro-mechanical coupling enhancers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7P7Y
 
 | | X-ray structure of Lactobacillus kefir alcohol dehydrogenase mutant Q126K | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bischoff, D, Walla, B, Janowski, R, Niessing, D, Weuster-Botz, D. | | Deposit date: | 2021-07-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Transfer of a Rational Crystal Contact Engineering Strategy between Diverse Alcohol Dehydrogenases
Crystals, 11, 2021
|
|
7XNN
 
 | | human KCNQ1-CaM-ML277-PIP2 complex in state B | | Descriptor: | (2R)-N-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-1-(4-methylbenzene-1-sulfonyl)piperidine-2-carboxamide, Calmodulin-3, POTASSIUM ION, ... | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural mechanisms for the activation of human cardiac KCNQ1 channel by electro-mechanical coupling enhancers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1VTD
 
 | | UNUSUAL HELICAL PACKING IN CRYSTALS OF DNA BEARING A MUTATION HOT SPOT | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*GP*CP*GP*CP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*GP*CP*GP*CP*CP*GP*GP*T)-3') | | Authors: | Timsit, Y, Westhof, E, Fuchs, R.P.P, Moras, D. | | Deposit date: | 1996-12-12 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unusual helical packing in crystals of DNA bearing a mutation hot spot.
Nature, 341, 1989
|
|
4E0M
 
 | | SVQIVYK segment from human Tau (305-311) displayed on 54-membered macrocycle scaffold (form I) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide SVQIVYK(ORN)EF(HAO)(4BF)K(ORN), PHOSPHATE ION | | Authors: | Zhao, M, Liu, C, Michael, S.R, Eisenberg, D. | | Deposit date: | 2012-03-04 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Out-of-register beta-sheets suggest a pathway
to toxic amyloid aggregates
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
