4XT3
 
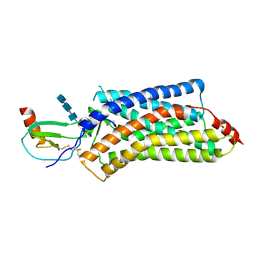 | | Structure of a viral GPCR bound to human chemokine CX3CL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fractalkine, G-protein coupled receptor homolog US28, ... | | Authors: | Burg, J.S, Jude, K.M, Waghray, D, Garcia, K.C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor.
Science, 347, 2015
|
|
6PGI
 
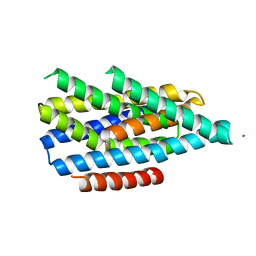 | | Asymmetric functions of a binuclear metal cluster within the transport pathway of the ZIP transition metal transporters | | Descriptor: | BbZIP, CADMIUM ION | | Authors: | Zhang, T, Sui, D, Zhang, C, Logan, T, Hu, J. | | Deposit date: | 2019-06-24 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Asymmetric functions of a binuclear metal center within the transport pathway of a human zinc transporter ZIP4.
Faseb J., 34, 2020
|
|
1ESB
 
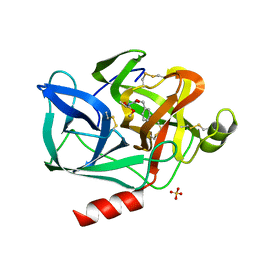 | | DIRECT STRUCTURE OBSERVATION OF AN ACYL-ENZYME INTERMEDIATE IN THE HYDROLYSIS OF AN ESTER SUBSTRATE BY ELASTASE | | Descriptor: | CALCIUM ION, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, PORCINE PANCREATIC ELASTASE, ... | | Authors: | Ding, X, Rasmussen, B, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-04 | | Release date: | 1994-04-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Direct structural observation of an acyl-enzyme intermediate in the hydrolysis of an ester substrate by elastase.
Biochemistry, 33, 1994
|
|
3B8W
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221P | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3B2C
 
 | | Crystal structure of the collagen triple helix model [{PRO-HYP(R)-GLY}4-{HYP(S)-Pro-GLY}2-{PRO-HYP(R)-GLY}4]3 | | Descriptor: | Collagen-like peptide | | Authors: | Motooka, D, Kawahara, K, Nakamura, S, Doi, M, Nishi, Y, Nishiuchi, Y, Nakazawa, T, Yoshida, T, Ohkubo, T, Kobayashi, Y, Kang, Y.K, Uchiyama, S. | | Deposit date: | 2011-07-26 | | Release date: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The triple helical structure and stability of collagen model peptide with 4(S)-hydroxyprolyl-pro-gly units
Biopolymers, 98, 2011
|
|
2VTH
 
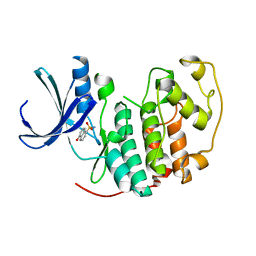 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H-pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design | | Descriptor: | 5-hydroxynaphthalene-1-sulfonamide, CELL DIVISION PROTEIN KINASE 2, GLYCEROL | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2VTP
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, {[(2,6-difluorophenyl)carbonyl]amino}-N-(4-fluorophenyl)-1H-pyrazole-3-carboxamide | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
6VRS
 
 | | Single particle reconstruction of glucose isomerase from Streptomyces rubiginosus based on data acquired in the presence of substantial aberrations | | Descriptor: | MANGANESE (II) ION, xylose isomerase | | Authors: | Bromberg, R, Guo, Y, Borek, D, Otwinowski, Z. | | Deposit date: | 2020-02-09 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | High-resolution cryo-EM reconstructions in the presence of substantial aberrations
Iucrj, 7, 2020
|
|
6VSC
 
 | |
3BB6
 
 | | Crystal structure of the P64488 protein from E.coli (strain K12). Northeast Structural Genomics Consortium target ER596 | | Descriptor: | Uncharacterized protein yeaR, ZINC ION | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Wang, D, Janjua, H, Owens, L, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray structure of the P64488 from E.coli (strain K12).
To be Published
|
|
3BDR
 
 | | Crystal structure of fatty acid-binding protein-like Ycf58 from Thermosynecoccus elongatus. Northeast Structural Genomics Consortium target TeR13. | | Descriptor: | PHOSPHATE ION, Ycf58 protein | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Forouhar, F, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-15 | | Release date: | 2007-11-27 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of fatty acid-binding protein-like Ycf58 from Thermosynecoccus elongatus.
To be Published
|
|
3B3C
 
 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine phosphonic acid | | Descriptor: | Bacterial leucyl aminopeptidase, LEUCINE PHOSPHONIC ACID, POTASSIUM ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-19 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
4XI0
 
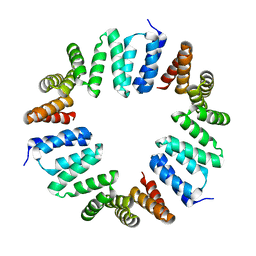 | | MamA 41-end from Desulfovibrio magneticus RS-1 | | Descriptor: | Magnetosome protein MamA | | Authors: | Zarivach, R, Zeytuni, N, Cronin, S, Davidov, G, Baran, D, Stein, T. | | Deposit date: | 2015-01-06 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | MamA as a Model Protein for Structure-Based Insight into the Evolutionary Origins of Magnetotactic Bacteria.
Plos One, 10, 2015
|
|
6VMG
 
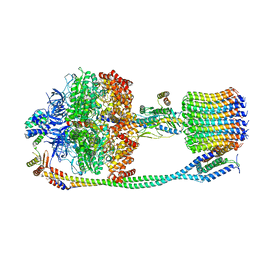 | | Chloroplast ATP synthase (O3, CF1FO) | | Descriptor: | ATP synthase delta chain, chloroplastic, ATP synthase epsilon chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.46 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
3BEW
 
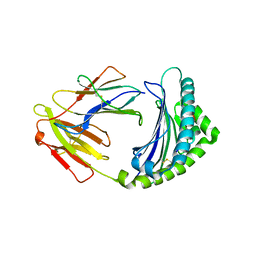 | | 10mer Crystal Structure of chicken MHC class I haplotype B21 | | Descriptor: | 10-mer from Tubulin beta-6 chain, Beta-2-microglobulin, Major histocompatibility complex class I glycoprotein haplotype B21 | | Authors: | Koch, M, Camp, S, Collen, T, Avila, D, Salomonsen, J, Wallny, H.J, van Hateren, A, Hunt, L, Jacob, J.P, Johnston, F, Marston, D.A, Shaw, I, Dunbar, P.R, Cerundolo, V, Jones, E.Y, Kaufman, J. | | Deposit date: | 2007-11-20 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of an MHC class I molecule from b21 chickens illustrate promiscuous Peptide binding
Immunity, 27, 2007
|
|
4G76
 
 | |
3B75
 
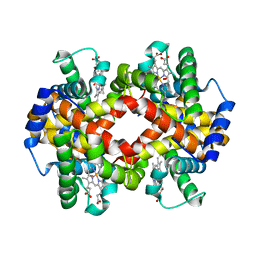 | | Crystal Structure of Glycated Human Haemoglobin | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Saraswathi, N.T, Syakhovich, V.E, Bokut, S.B, Moras, D, Ruff, M. | | Deposit date: | 2007-10-30 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The effect of hemoglobin glycosylation on diabete linked oxidative stress
To be Published
|
|
6PRX
 
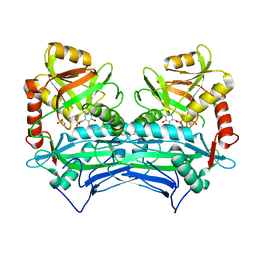 | | oxidized Human Branched Chain Aminotransferase mutant C318A | | Descriptor: | Branched-chain-amino-acid aminotransferase, mitochondrial, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dong, M, Herbert, D, Gibbs, S. | | Deposit date: | 2019-07-11 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of an oxidized mutant of human mitochondrial branched-chain aminotransferase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
3B9C
 
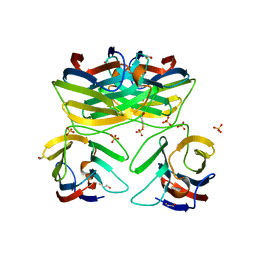 | | Crystal Structure of Human GRP CRD | | Descriptor: | BETA-MERCAPTOETHANOL, HSPC159, SULFATE ION | | Authors: | Zhou, D, Ge, H.H, Niu, L.W, Teng, M.K. | | Deposit date: | 2007-11-05 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the C-terminal conserved domain of human GRP, a galectin-related protein, reveals a function mode different from those of galectins.
Proteins, 71, 2008
|
|
3B3S
 
 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine | | Descriptor: | Bacterial leucyl aminopeptidase, LEUCINE, SODIUM ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-22 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
4GKR
 
 | |
6P7E
 
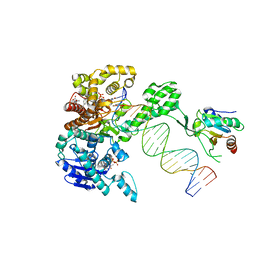 | | Structure of T7 DNA Polymerase Bound to a Primer/Template DNA and a Peptide that Mimics the C-terminal Tail of the Primase-Helicase | | Descriptor: | ASP-THR-ASP-PHE peptide, DNA (25-MER), DNA (5'-D(P*GP*GP*CP*AP*GP*GP*TP*GP*GP*TP*CP*TP*TP*GP*CP*CP*GP*GP*TP*GP*A)-3'), ... | | Authors: | Foster, B.M, Rosenberg, D, Salvo, H, Stephens, K.L, Bintz, B.J, Hammel, M, Ellenberger, T, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2019-06-05 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Combined Solution and Crystal Methods Reveal the Electrostatic Tethers That Provide a Flexible Platform for Replication Activities in the Bacteriophage T7 Replisome.
Biochemistry, 58, 2019
|
|
6PJJ
 
 | | Human PRPF4B bound to benzothiophene inhibitor 224 | | Descriptor: | 1,2-ETHANEDIOL, 4-(5-{[(3-aminophenyl)methyl]carbamoyl}thiophen-2-yl)-1-benzothiophene-2-carboxamide, PHOSPHATE ION, ... | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-28 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | to be published
To Be Published
|
|
4R5N
 
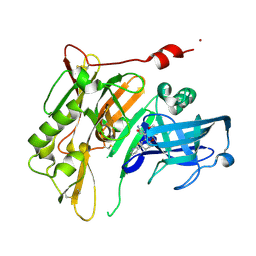 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | Descriptor: | (4R,4a'S,10a'R)-8'-(2-fluoropyridin-3-yl)-4a'-methyl-3',4',4a',10a'-tetrahydro-2'H-spiro[1,3-oxazole-4,10'-pyrano[3,2-b]chromen]-2-amine, Beta-secretase 1, NICKEL (II) ION | | Authors: | Vigers, G.P.A, Smith, D. | | Deposit date: | 2014-08-21 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
6P7X
 
 | | Structure of the K. lactis CBF3 core - Ndc10 D1D2 complex | | Descriptor: | Cep3, Ctf13, Ndc10, ... | | Authors: | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
