7MNG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor VBY-825 (Partial Occupancy) | | 分子名称: | (2R,3S)-N-cyclopropyl-3-{[(2R)-3-(cyclopropylmethanesulfonyl)-2-{[(1S)-2,2,2-trifluoro-1-(4-fluorophenyl)ethyl]amino}propanoyl]amino}-2-hydroxypentanamide (non-preferred name), 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2021-04-30 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7MHJ
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 298 K and High Humidity | | 分子名称: | 3C-like proteinase, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.0005 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHO
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 298 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
7MHH
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 277 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1908 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHI
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 298 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHQ
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 310 K | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9601 Å) | | 主引用文献 | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
7MHP
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 298 K at high humidity | | 分子名称: | 3C-like proteinase, ZINC ION | | 著者 | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | 登録日 | 2021-04-15 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.0005 Å) | | 主引用文献 | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
3TH2
 
 | | Mg2+ Is Required for Optimal Folding of the Gamma-Carboxyglutamic Acid (Gla) Domains of Vitamin K-Dependent Clotting Factors At Physiological Ca2+ | | 分子名称: | BENZAMIDINE, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Vadivel, K, Agah, S, Cascio, D, Padmanabhan, K, Bajaj, S.P. | | 登録日 | 2011-08-18 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Mg2+ Is Required for Optimal Folding of the Gamma-Carboxyglutamic Acid (Gla)
Domains of Vitamin K-Dependent Clotting Factors At Physiological Ca2+
To be Published
|
|
3TBW
 
 | | CRYSTAL STRUCTURE OF THE MURINE CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX H-2DB IN COMPLEX WITH THE LCMV-DERIVED GP33 ALTERED PEPTIDE ligand (A2G, V3P, Y4S) | | 分子名称: | Beta-2-microglobulin, GLYCOPROTEIN GPC, H-2 class I histocompatibility antigen, ... | | 著者 | Duru, A.D, Allerbring, E.B, Uchtenhagen, H, Mazumdar, P.A, Badia-Martinez, D, Madhurantakam, C, Sandalova, T, Nygren, P, Achour, A. | | 登録日 | 2011-08-08 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Conversion of a T cell viral antagonist into an agonist through higher stabilization and conserved molecular mimicry: Implications for TCR recognition
To be Published
|
|
7KQY
 
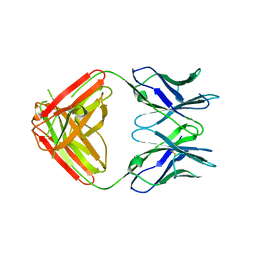 | | Crystal Structure and Characterization of Human Heavy-Chain only Antibodies reveals a novel, stable dimeric structure similar to Monoclonal Antibodies | | 分子名称: | Heavy-Chain only Human Antibodies | | 著者 | Bahmanjah, S, Mieczkowski, C, Yu, Y, Baker, J, Raghunathan, G, Tomazela, D, Hsieh, M, Mccoy, M, Strickland, C, Fayadat-Dilman, L. | | 登録日 | 2020-11-18 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.913 Å) | | 主引用文献 | Crystal Structure and Characterization of Human Heavy-Chain Only Antibodies Reveals a Novel, Stable Dimeric Structure Similar to Monoclonal Antibodies.
Antibodies, 9, 2020
|
|
3TG2
 
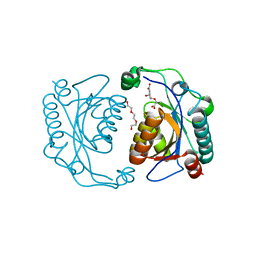 | | Crystal structure of the ISC domain of VibB in complex with isochorismate | | 分子名称: | (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, TRIETHYLENE GLYCOL, Vibriobactin-specific isochorismatase | | 著者 | Liu, S, Zhang, C, Niu, B, Li, N, Liu, X, Liu, M, Wei, T, Zhu, D, Huang, Y, Xu, S, Gu, L. | | 登録日 | 2011-08-17 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.101 Å) | | 主引用文献 | Structural insight into the ISC domain of VibB from Vibrio cholerae at atomic resolution: a snapshot just before the enzymatic reaction
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7KIP
 
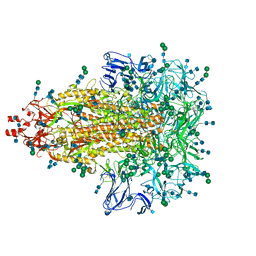 | | A 3.4 Angstrom cryo-EM structure of the human coronavirus spike trimer computationally derived from vitrified NL63 virus particles | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Zhang, K, Li, S, Pintilie, G, Chmielewski, D, Schmid, M, Simmons, G, Jin, J, Chiu, W. | | 登録日 | 2020-10-24 | | 公開日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | A 3.4- angstrom cryo-EM structure of the human coronavirus spike trimer computationally derived from vitrified NL63 virus particles.
Biorxiv, 2020
|
|
7KVA
 
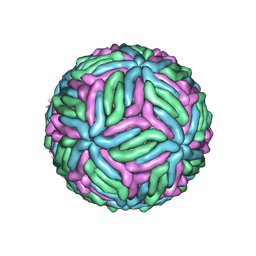 | | Structure of West Nile virus (Kunjin) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Matrix protein M | | 著者 | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | 登録日 | 2020-11-27 | | 公開日 | 2020-12-23 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses.
Nat Commun, 12, 2021
|
|
7KV9
 
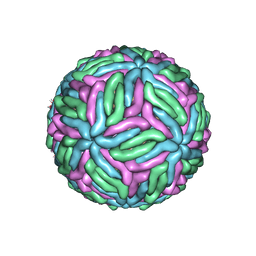 | | Chimeric flavivirus between Binjari virus and West Nile (Kunjin) virus | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Matrix protein M, envelope protein E | | 著者 | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | 登録日 | 2020-11-27 | | 公開日 | 2020-12-23 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses.
Nat Commun, 12, 2021
|
|
7KWJ
 
 | | Spermidine N-acetyltransferase SpeG K23-Q34 chimera from Vibrio cholerae and hSSAT | | 分子名称: | Spermidine N(1)-acetyltransferase | | 著者 | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K, Kuhn, M.L. | | 登録日 | 2020-12-01 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
3U26
 
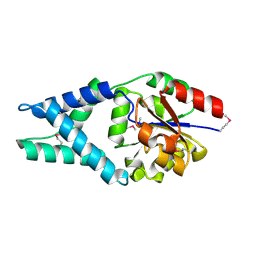 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR48 | | 分子名称: | PF00702 domain protein | | 著者 | Seetharaman, J, Lew, S, Nivon, L, Baker, D, Bjelic, S, Ciccosanti, C, Sahdev, S, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-09-30 | | 公開日 | 2011-11-23 | | 最終更新日 | 2022-03-02 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Computational design of enone-binding proteins with catalytic activity for the Morita-Baylis-Hillman reaction.
Acs Chem.Biol., 8, 2013
|
|
3U2Y
 
 | | ATP synthase c10 ring in proton-unlocked conformation at pH 6.1 | | 分子名称: | ATP synthase subunit C, mitochondrial | | 著者 | Symersky, J, Pagadala, V, Osowski, D, Krah, A, Meier, T, Faraldo-Gomez, J, Mueller, D.M. | | 登録日 | 2011-10-04 | | 公開日 | 2012-02-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of the c(10) ring of the yeast mitochondrial ATP synthase in the open conformation.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3TKZ
 
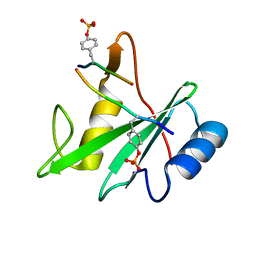 | | Structure of the SHP-2 N-SH2 domain in a 1:2 complex with RVIpYFVPLNR peptide | | 分子名称: | PROTEIN (RVIpYFVPLNR peptide), Tyrosine-protein phosphatase non-receptor type 11 | | 著者 | Zhang, Y, Zhang, J, Yuan, C, Hard, R.L, Park, I.H, Li, C, Bell, C.E, Pei, D. | | 登録日 | 2011-08-29 | | 公開日 | 2011-10-26 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Simultaneous binding of two peptidyl ligands by a SRC homology 2 domain.
Biochemistry, 50, 2011
|
|
7KV8
 
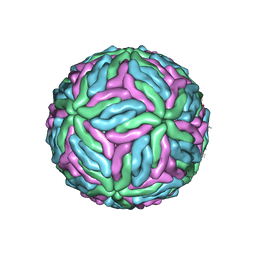 | | Chimeric flavivirus between Binjari virus and Dengue virus serotype-2 | | 分子名称: | (2S,3R,4Z)-3-hydroxy-2-[(9E)-octadec-9-enoylamino]octadec-4-en-1-yl dihydrogen phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | 著者 | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | 登録日 | 2020-11-27 | | 公開日 | 2020-12-23 | | 最終更新日 | 2021-11-24 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses.
Nat Commun, 12, 2021
|
|
7MRR
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Leupeptin | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, LEUPEPTIN | | 著者 | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2021-05-08 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7MQV
 
 | | Crystal structure of truncated (ACT domain removed) prephenate dehydrogenase tyrA from Bacillus anthracis in complex with NAD | | 分子名称: | CHLORIDE ION, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Shabalin, I.G, Gritsunov, A, Gabryelska, A, Czub, M.P, Grabowski, M, Cooper, D.R, Christendat, D, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-05-06 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The crystal structure of Bacillus anthracis prephenate dehydrogenase identified an ACT regulatory domain and a novel mode of metabolic regulation for proteins within the prephenate dehydrogenase family of enzyme
to be published
|
|
7M53
 
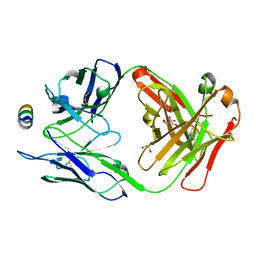 | | B6 Fab fragment bound to the SARS-CoV/SARS-CoV-2 spike stem helix peptide | | 分子名称: | B6 antigen-binding (Fab) fragment heavy chain, B6 antigen-binding (Fab) fragment light chain, GLYCEROL, ... | | 著者 | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2021-03-22 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M51
 
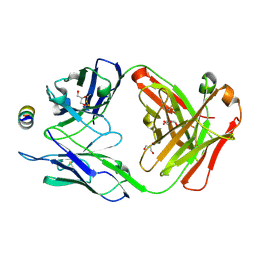 | | B6 Fab fragment bound to the OC43 spike stem helix peptide | | 分子名称: | B6 antigen-binding (Fab) fragment heavy chain, B6 antigen-binding (Fab) fragment light chain, GLYCEROL, ... | | 著者 | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2021-03-22 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M55
 
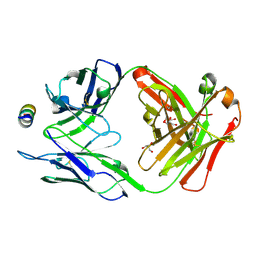 | | B6 Fab fragment bound to the MERS-CoV spike stem helix peptide | | 分子名称: | B6 antigen binding fragment (Fab) heavy chain, B6 antigen binding fragment (Fab) light chain, GLYCEROL, ... | | 著者 | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2021-03-22 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M5E
 
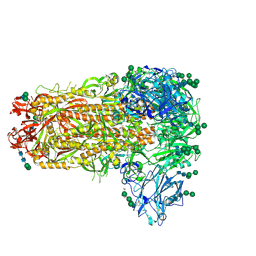 | |
