1NJ5
 
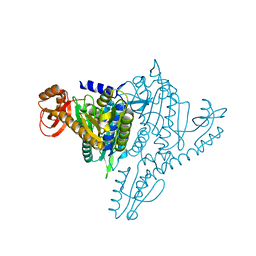 | | Crystal structure of Prolyl-tRNA Synthetase from Methanothermobacter thermautotrophicus bound to proline sulfamoyl adenylate | | 分子名称: | '5'-O-(N-(L-PROLYL)-SULFAMOYL)ADENOSINE, MAGNESIUM ION, Proline-tRNA Synthetase, ... | | 著者 | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | 登録日 | 2002-12-30 | | 公開日 | 2003-03-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7MZ6
 
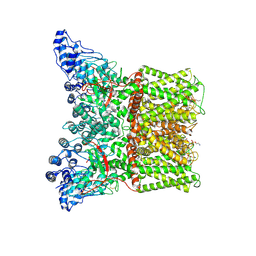 | | Cryo-EM structure of minimal TRPV1 with 1 perturbed PI | | 分子名称: | (10R,13S)-16-amino-13-hydroxy-7,13-dioxo-8,12,14-trioxa-13lambda~5~-phosphahexadecan-10-yl hexadecanoate, (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, ... | | 著者 | Zhang, K, Julius, D, Cheng, Y. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-22 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZB
 
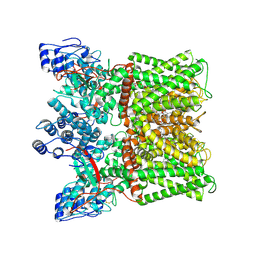 | | Cryo-EM structure of minimal TRPV1 with 3 bound RTX and 1 perturbed PI | | 分子名称: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | 著者 | Zhang, K, Julius, D, Cheng, Y. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-22 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.72 Å) | | 主引用文献 | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZC
 
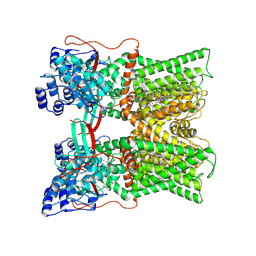 | |
2RO2
 
 | | Solution structure of domain I of the negative polarity CChMVd hammerhead ribozyme | | 分子名称: | RNA (5'-R(*GP*GP*GP*AP*GP*AP*CP*CP*UP*GP*AP*AP*GP*UP*GP*GP*GP*UP*UP*UP*CP*CP*C)-3') | | 著者 | Gallego, J, Dufour, D, Gago, S, de la Pena, M, Flores, R. | | 登録日 | 2008-03-05 | | 公開日 | 2008-12-30 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-function analysis of the ribozymes of chrysanthemum chlorotic mottle viroid: a loop-loop interaction motif conserved in most natural hammerheads
Nucleic Acids Res., 37, 2009
|
|
7MZA
 
 | | Cryo-EM structure of minimal TRPV1 with 2 bound RTX in adjacent pockets | | 分子名称: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | 著者 | Zhang, K, Julius, D, Cheng, Y. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-22 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZ9
 
 | | Cryo-EM structure of minimal TRPV1 with 1 partially bound RTX | | 分子名称: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | 著者 | Zhang, K, Julius, D, Cheng, Y. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-22 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZ7
 
 | |
7MZD
 
 | |
2RV8
 
 | |
7MZE
 
 | | Cryo-EM structure of minimal TRPV1 with 2 bound RTX in opposite pockets | | 分子名称: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | 著者 | Zhang, K, Julius, D, Cheng, Y. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-22 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7NKZ
 
 | | Cryo-EM structure of the cytochrome bd oxidase from M. tuberculosis at 2.5 A resolution | | 分子名称: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, HEME B/C, MENAQUINONE-9, ... | | 著者 | Safarian, S, Wu, D, Krause, K.L, Michel, H. | | 登録日 | 2021-02-19 | | 公開日 | 2021-09-22 | | 最終更新日 | 2021-09-29 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | The cryo-EM structure of the bd oxidase from M. tuberculosis reveals a unique structural framework and enables rational drug design to combat TB.
Nat Commun, 12, 2021
|
|
1NE2
 
 | | Crystal Structure of Thermoplasma acidophilum 1320 (APC5513) | | 分子名称: | FORMIC ACID, hypothetical protein ta1320 | | 著者 | Kim, Y, Joachimiak, A, Edwards, A, Xu, X, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-12-10 | | 公開日 | 2003-07-01 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structure of Thermoplasma acidophilum 1320 (APC5513)
To be Published
|
|
7ZGV
 
 | | Serratia NucC bound to cA3 | | 分子名称: | ACETATE ION, CALCIUM ION, RNA (5'-R(P*AP*AP*A)-3'), ... | | 著者 | Garcia-Doval, C, Mayo-Munoz, D, Smith, L.M, Fineran, P.C. | | 登録日 | 2022-04-04 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Type III CRISPR-Cas provides resistance against nucleus-forming jumbo phages via abortive infection.
Mol.Cell, 82, 2022
|
|
7ZGW
 
 | |
2RMB
 
 | |
1NIX
 
 | |
8A5Q
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 on straight DNA | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin related protein 4 (Arp4), ... | | 著者 | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | 登録日 | 2022-06-15 | | 公開日 | 2022-12-14 | | 最終更新日 | 2022-12-28 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
1NJ8
 
 | | Crystal Structure of Prolyl-tRNA Synthetase from Methanocaldococcus janaschii | | 分子名称: | Proline-tRNA Synthetase | | 著者 | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | 登録日 | 2002-12-30 | | 公開日 | 2003-03-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7NHG
 
 | |
8A5P
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 on curved DNA | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin related protein 4 (Arp4), ... | | 著者 | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | 登録日 | 2022-06-15 | | 公開日 | 2022-12-14 | | 最終更新日 | 2022-12-28 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8A5A
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of INO80 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-like protein ARP8, ... | | 著者 | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | 登録日 | 2022-06-14 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
7NIA
 
 | |
4HN4
 
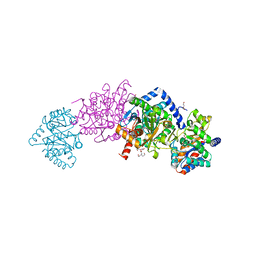 | | Tryptophan synthase in complex with alpha aminoacrylate E(A-A) form and the F9 inhibitor in the alpha site | | 分子名称: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, BICINE, ... | | 著者 | Hilario, E, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | 登録日 | 2012-10-18 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states.
Biochemistry, 52, 2013
|
|
7NI9
 
 | |
