5QOI
 
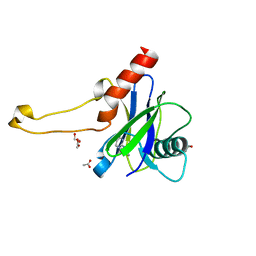 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with FMOPL000213a | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QP9
 
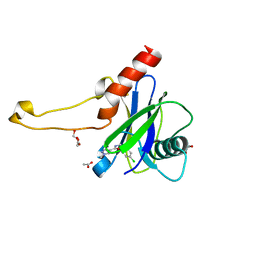 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z100435060 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5UDG
 
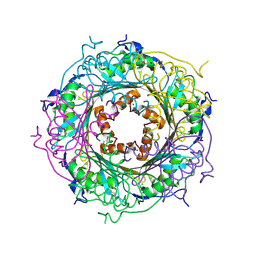 | | Mutant E97Q crystal structure of Bacillus subtilis QueF with a disulfide Cys 55-99 | | Descriptor: | MAGNESIUM ION, NADPH-dependent 7-cyano-7-deazaguanine reductase, TRIETHYLENE GLYCOL | | Authors: | Mohammad, A, Kiani, M.K, Iwata-Reuyl, D, Stec, B, Swairjo, M. | | Deposit date: | 2016-12-27 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protection of the Queuosine Biosynthesis Enzyme QueF from Irreversible Oxidation by a Conserved Intramolecular Disulfide.
Biomolecules, 7, 2017
|
|
5QP1
 
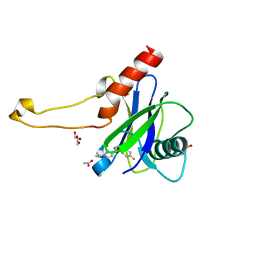 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z1190363272 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QOJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with FMOCR000171b | | Descriptor: | 1,2-ETHANEDIOL, 2-(thiophen-2-yl)-1H-imidazole, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QP3
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z1494850193 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5TIC
 
 | | X-ray structure of wild-type E. coli Acyl-CoA thioesterase I at pH 5 | | Descriptor: | Acyl-CoA thioesterase I, CHLORIDE ION | | Authors: | Thoden, J.B, Holden, H.M, Grisewood, M.J, Hernandez Lozada, N.J, Gifford, N.P, Mendez-Perez, D, Schoenberger, H.A, Allan, M.F, Pfleger, B.F, Marines, C.D. | | Deposit date: | 2016-10-02 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Computational Redesign of Acyl-ACP Thioesterase with Improved Selectivity toward Medium-Chain-Length Fatty Acids.
ACS Catal, 7, 2017
|
|
5TJ3
 
 | | Crystal structure of wild type alkaline phosphatase PafA to 1.7A resolution | | Descriptor: | Alkaline phosphatase PafA, ZINC ION | | Authors: | Lyubimov, A.Y, Sunden, F, Ressl, S, Herschlag, D. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanistic and Evolutionary Insights from Comparative Enzymology of Phosphomonoesterases and Phosphodiesterases across the Alkaline Phosphatase Superfamily.
J.Am.Chem.Soc., 138, 2016
|
|
5NBY
 
 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, FLAVIN MONONUCLEOTIDE | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2017-03-02 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
5N5S
 
 | | Crystal structure of aldehyde dehydrogenase 21 (ALDH21) from Physcomitrella patens in complex with NADP+ | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase 21 (ALDH21), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kopecny, D, Vigouroux, A, Briozzo, P, Morera, S. | | Deposit date: | 2017-02-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The ALDH21 gene found in lower plants and some vascular plants codes for a NADP(+) -dependent succinic semialdehyde dehydrogenase.
Plant J., 92, 2017
|
|
5N6Q
 
 | | Xenobiotic reductase A (XenA) from Pseudomonas putida in complex with 2-phenylacrylic acid | | Descriptor: | 2-Phenylacrylic acid, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the ene-reductase synthesis of profens.
Org. Biomol. Chem., 15, 2017
|
|
5NAP
 
 | | Torpedo californica acetylcholinesterase in complex with a non-chiral donepezil-like inhibitor 17 | | Descriptor: | (2~{E})-5,6-dimethoxy-2-[[1-(phenylmethyl)piperidin-4-yl]methylidene]-3~{H}-inden-1-one, Acetylcholinesterase, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Caliandro, R, Pesaresi, A, Lamba, D. | | Deposit date: | 2017-02-28 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Kinetic and structural studies on the interactions of Torpedo californica acetylcholinesterase with two donepezil-like rigid analogues.
J Enzyme Inhib Med Chem, 33, 2018
|
|
5NG1
 
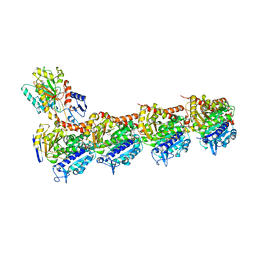 | | TUBULIN-MTC-zampanolide complex | | Descriptor: | (2Z,4E)-N-[(S)-[(1S,2E,5S,8E,10Z,17S)-3,11-dimethyl-19-methylidene-7,13-dioxo-6,21-dioxabicyclo[15.3.1]henicosa-2,8,10-trien-5-yl](hydroxy)methyl]hexa-2,4-dienamide, (2~{Z},4~{E})-~{N}-[(~{S})-oxidanyl-[(1~{S},2~{E},5~{S},11~{R},17~{S},19~{R})-3,11,19-trimethyl-7,13-bis(oxidanylidene)-6,21-dioxabicyclo[15.3.1]henicos-2-en-5-yl]methyl]hexa-2,4-dienamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Field, J.J, Pera, B, Estevez Gallego, J, Calvo, E, Rodriguez-Salarichs, J, Saez-Calvo, G, Zuwerra, D, Jordi, M, Prota, A.E, Menchon, G, Miller, J.H, Altmann, K.-H, Diaz, J.F. | | Deposit date: | 2017-03-16 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zampanolide Binding to Tubulin Indicates Cross-Talk of Taxane Site with Colchicine and Nucleotide Sites.
J. Nat. Prod., 81, 2018
|
|
5T8C
 
 | |
5NK7
 
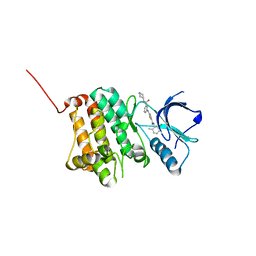 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2a | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-[[(3~{R},4~{R})-3-fluoranylpiperidin-4-yl]carbamoyl]phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NKF
 
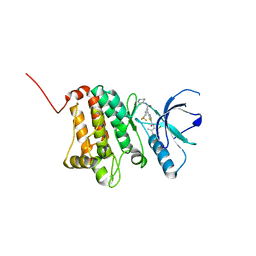 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 3b | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-(piperidin-4-ylcarbamoyl)-5-(trifluoromethyl)phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NK3
 
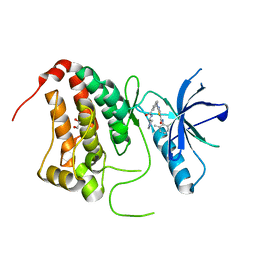 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 1l | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-[[(3~{S})-pyrrolidin-3-yl]carbamoyl]phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.586 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NKC
 
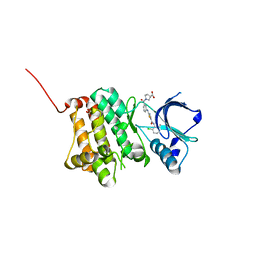 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2h | | Descriptor: | (3~{S})-1-[3-[[5-[(2-chloranyl-6-methyl-phenyl)carbamoyl]-1,3-thiazol-2-yl]amino]phenyl]carbonylpyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NP1
 
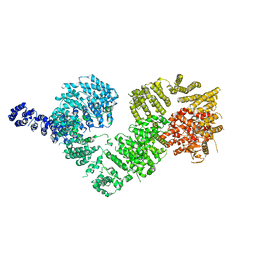 | | Open protomer of human ATM (Ataxia telangiectasia mutated) | | Descriptor: | Serine-protein kinase ATM | | Authors: | Baretic, D, Pollard, H.K, Fisher, D.I, Johnson, C.M, Santhanam, B, Truman, C.M, Kouba, T, Fersht, A.R, Phillips, C, Williams, R.L. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of closed and open conformations of dimeric human ATM.
Sci Adv, 3, 2017
|
|
5NQK
 
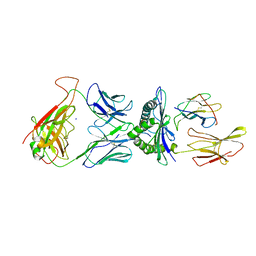 | | human 199.16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Exertier, C, Reiser, J.-B, Lantez, V, Chouquet, A, Bonneville, M, Saulquin, X, Housset, D. | | Deposit date: | 2017-04-20 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | human 199.16 TCR in complex with Melan-A/MART-1 (26-35) peptide and HLA-A2
To Be Published
|
|
5NR5
 
 | |
5T1A
 
 | | Structure of CC Chemokine Receptor 2 with Orthosteric and Allosteric Antagonists | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{R})-1-(4-chloranyl-2-fluoranyl-phenyl)-2-cyclohexyl-3-ethanoyl-4-oxidanyl-2~{H}-pyrrol-5-one, (3S)-1-{(1S,2R,4R)-4-[methyl(propan-2-yl)amino]-2-propylcyclohexyl}-3-{[6-(trifluoromethyl)quinazolin-4-yl]amino}pyrrolidin-2-one, ... | | Authors: | Zheng, Y, Qin, L, Ortiz Zacarias, N.V, de Vries, H, Han, G.W, Gustavsson, M, Dabros, M, Zhao, C, Cherney, R.J, Carter, P, Stamos, D, Abagyan, R, Cherezov, V, Stevens, R.C, IJzerman, A.P, Heitman, L.H, Tebben, A, Kufareva, I, Handel, T.M. | | Deposit date: | 2016-08-18 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists.
Nature, 540, 2016
|
|
5SY7
 
 | | Crystal Structure of the Heterodimeric NPAS3-ARNT Complex with HRE DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(*CP*AP*CP*GP*AP*CP*CP*CP*GP*CP*AP*CP*GP*TP*AP*CP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*GP*CP*GP*TP*AP*CP*GP*TP*GP*CP*GP*GP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Wu, D, Su, X, Potluri, N, Kim, Y, Rastinejad, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors.
Elife, 5, 2016
|
|
5NRE
 
 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - a3GalT in complex with lactose - a3GalT-LAT | | Descriptor: | N-acetyllactosaminide alpha-1,3-galactosyltransferase, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5UTL
 
 | |
