6GH4
 
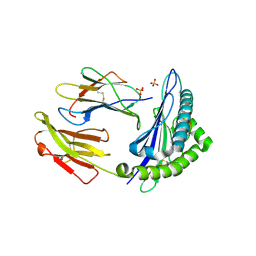 | | HLA-E*01:03 in complex with the Mtb44 peptide variant: Mtb44*P2-Gln. | | Descriptor: | ARG-GLN-PRO-ALA-LYS-ALA-PRO-LEU-LEU, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | Deposit date: | 2018-05-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
5EFH
 
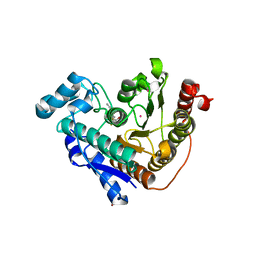 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 in complex with trifluoroketone transition state analogue | | Descriptor: | 1,2-ETHANEDIOL, 7-[(3-aminopropyl)amino]-1,1,1-trifluoroheptane-2,2-diol, BROMIDE ION, ... | | Authors: | Hai, Y, Christianson, D.W. | | Deposit date: | 2015-10-23 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Histone deacetylase 6 structure and molecular basis of catalysis and inhibition.
Nat.Chem.Biol., 12, 2016
|
|
8G8B
 
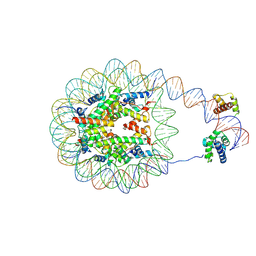 | | Nucleosome with human nMatn1 sequence in complex with Human Oct4 | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Sinha, K.K, Bilokapic, S, Du, Y, Malik, D, Halic, M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Histone modifications regulate pioneer transcription factor cooperativity.
Nature, 619, 2023
|
|
6THH
 
 | |
8G8G
 
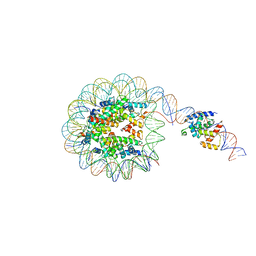 | | Interaction of H3 tail in LIN28B nucleosome with Oct4 | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Sinha, K.K, Bilokapic, S, Du, Y, Malik, D, Halic, M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-03-22 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Histone modifications regulate pioneer transcription factor cooperativity.
Nature, 619, 2023
|
|
8I6N
 
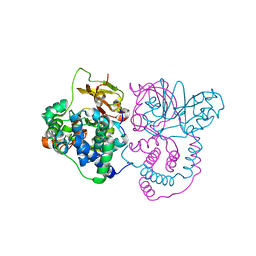 | | Crystal structure of Co-type nitrile hydratase mutant from Pseudomonas thermophila - L6T | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta | | Authors: | Ma, D, Cheng, Z.Y, Lai, Q.P, Hou, X.D, Yin, D.J, Rao, Y.J, Zhou, Z.M. | | Deposit date: | 2023-01-29 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Co-type nitrile hydratase mutant L6T from Pseudomonas thermophila at 2.2 Angstroms resolution.
To Be Published
|
|
6QG4
 
 | |
8GRM
 
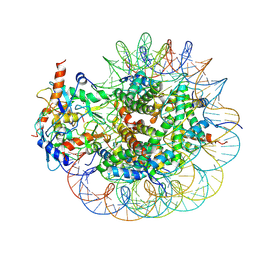 | | Cryo-EM structure of PRC1 bound to H2AK119-UbcH5b-Ub nucleosome | | Descriptor: | COMMD3 protein, DNA (144-MER), DNA (145-MER), ... | | Authors: | Ai, H.S, Zebin, T, Zhihend, D, Jiakun, T, Liying, Z, Jia-Bin, L, Man, P, Liu, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Synthetic E2-Ub-nucleosome conjugates for studying nucleosome ubiquitination.
Chem, 2023
|
|
6QHT
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 376 ms | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
8QOH
 
 | | Crystal structure of the kinetoplastid kinetochore protein KKT14 C-terminal domain from Apiculatamorpha spiralis | | Descriptor: | kinetochore protein KKT14 | | Authors: | Carter, W, Ballmer, D, Ishii, M, Ludzia, P, Akiyoshi, B. | | Deposit date: | 2023-09-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Kinetoplastid kinetochore proteins KKT14-KKT15 are divergent Bub1/BubR1-Bub3 proteins.
Open Biology, 14, 2024
|
|
6QI2
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 13536 ms | | Descriptor: | Fluoroacetate dehalogenase, GLYCOLIC ACID | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
8GVF
 
 | | The outward-facing structure of hAE2 in basic pH | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
7AX0
 
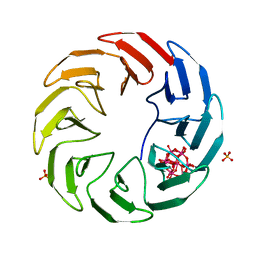 | | Crystal structure of the computationally designed Scone-E protein co-crystallized with STA form a | | Descriptor: | Keggin (STA), PHOSPHATE ION, SconeE | | Authors: | Mylemans, B, Vandebroek, L, Parac-Vogt, T.N, Voet, A.R.D. | | Deposit date: | 2020-11-09 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Scone: pseudosymmetric folding of a symmetric designer protein.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4YVA
 
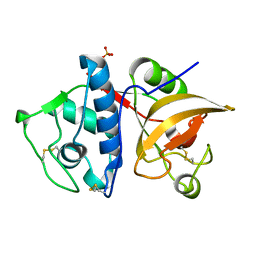 | | Cathepsin K co-crystallized with actinomycetes extract | | Descriptor: | Cathepsin K, SULFATE ION | | Authors: | Aguda, A.H, Nguyen, N.T, Bromme, D, Brayer, G.D. | | Deposit date: | 2015-03-19 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Affinity Crystallography: A New Approach to Extracting High-Affinity Enzyme Inhibitors from Natural Extracts.
J.Nat.Prod., 79, 2016
|
|
8EV4
 
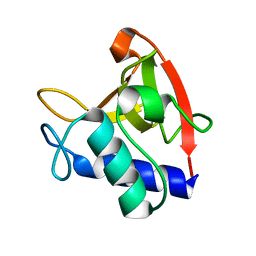 | | NlpC B3 - Trichomonas Vaginalis | | Descriptor: | Clan CA, family C40, NlpC/P60 superfamily cysteine peptidase | | Authors: | Barnett, M.J, Goldstone, D.C. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | NlpC/P60 peptidoglycan hydrolases of Trichomonas vaginalis have complementary activities that empower the protozoan to control host-protective lactobacilli.
Plos Pathog., 19, 2023
|
|
6C6X
 
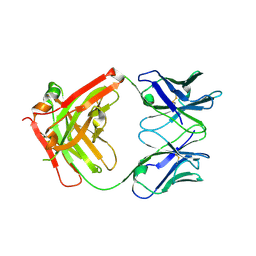 | | Crystal structure of Middle-East Respiratory Syndrome (MERS) coronavirus neutralizing antibody JC57-14 isolated from a vaccinated rhesus macaque. | | Descriptor: | JC57-14 Heavy chain, JC57-14 Light chain | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape.
J. Virol., 92, 2018
|
|
7AFL
 
 | | Bacterial 30S ribosomal subunit assembly complex state D (multibody refinement for body domain of 30S ribosome) | | Descriptor: | 16SrRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
4YSX
 
 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with the specific inhibitor NN23 | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
8IHG
 
 | |
8VEW
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA and oxamide compound 24 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 5-{2-[(2R,5S)-5-methyl-2-phenylpiperidin-1-yl](oxo)acetamido}pyridine-3-carboxamide, CHLORIDE ION, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
6Q6P
 
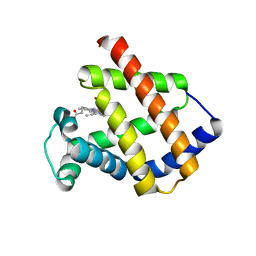 | | Antarctic fish cytoglobin 1 from D.mawsoni | | Descriptor: | Cytoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Giordano, D, Pesce, A, Vermeylen, S, Abbruzzetti, S, Nardini, M, Marchesani, F, Berghmans, H, Seira, C, Bruno, S, Luque, F.J, di Prisco, G, Ascenzi, P, Dewilde, S, Bolognesi, M, Viappiani, C, Verde, C. | | Deposit date: | 2018-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional properties of Antarctic fish cytoglobins-1: Cold-reactivity in multi-ligand reactions.
Comput Struct Biotechnol J, 18, 2020
|
|
7P2G
 
 | |
6VR5
 
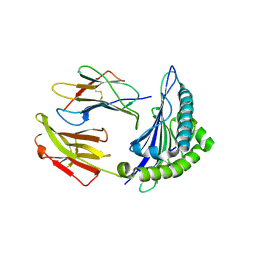 | | Complex of HLA-A2, a class I MHC, with a p53 peptide | | Descriptor: | Beta-2-microglobulin, Cellular tumor antigen p53 peptide, MHC class I antigen | | Authors: | Wu, D, Gallagher, D.T, Pierce, B.G, Mariuzza, R.A. | | Deposit date: | 2020-02-06 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
Nat Commun, 11, 2020
|
|
6QHU
 
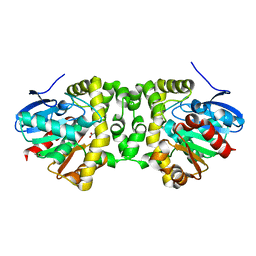 | | Time resolved structural analysis of the full turnover of an enzyme - 100 ms | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
6QI1
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 12312 ms | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
