5MZ7
 
 | | Crystal Structure of the third PDZ domain from the synaptic protein PSD-95 with incorporated Azidohomoalanine | | Descriptor: | Disks large homolog 4 | | Authors: | Kudlinzki, D, Lehner, F, Witt, K, Linhard, V.L, Silvers, R, Schwalbe, H. | | Deposit date: | 2017-01-31 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Impact of Azidohomoalanine Incorporation on Protein Structure and Ligand Binding.
Chembiochem, 18, 2017
|
|
4YAC
 
 | | Crystal structure of LigO in complex with NADH from Sphingobium sp. strain SYK-6 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C alpha-dehydrogenase | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural and Biochemical Characterization of the Early and Late Enzymes in the Lignin beta-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6.
J.Biol.Chem., 291, 2016
|
|
3G2J
 
 | | Crystal structure of 1-(beta-D-glucopyranosyl)-4-substituted-1,2,3-triazoles in complex with glycogen phosphorylase | | Descriptor: | Glycogen phosphorylase, muscle form, N-(hydroxyacetyl)-beta-D-glucopyranosylamine | | Authors: | Chrysina, E.D, Bokor, E, Alexacou, K.-M, Charavgi, M.-D, Oikonomakos, G.N, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2009-01-31 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Amide-1,2,3-triazole bioisosterism: the glycogen phosphorylase case
Tetrahedron: Asymmetry, 20, 2009
|
|
6N3K
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 1 | | Descriptor: | Epoxide hydrolase, N-{cis-4-[(2,6-difluorophenyl)methoxy]cyclohexyl}-N'-(3-phenylpropyl)urea | | Authors: | de Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
6Q6G
 
 | | Cryo-EM structure of the APC/C-Cdc20-Cdk2-cyclinA2-Cks2 complex, the D1 box class | | Descriptor: | Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Zhang, S, Barford, D. | | Deposit date: | 2018-12-11 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cyclin A2 degradation during the spindle assembly checkpoint requires multiple binding modes to the APC/C.
Nat Commun, 10, 2019
|
|
5UOP
 
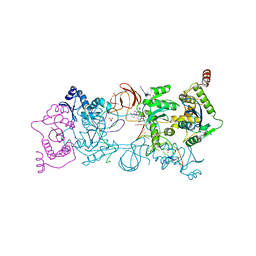 | | CRYSTAL STRUCTURE OF THE PROTOTYPE FOAMY VIRUS INTASOME WITH A 2- PYRIDINONE AMINAL INHIBITOR (COMPOUND 18) | | Descriptor: | (1S,2S,5R)-8'-[(3-chloro-4-fluorophenyl)methyl]-2'-[2-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)ethyl]-6'-hydroxy-9',10'-dihydro-2'H-spiro[bicyclo[3.1.0]hexane-2,3'-imidazo[5,1-a][2,6]naphthyridine]-1',5',7'(8'H)-trione, GLYCEROL, INTEGRASE, ... | | Authors: | Klein, D.J. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery and optimization of 2-pyridinone aminal integrase strand transfer inhibitors for the treatment of HIV.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6Q98
 
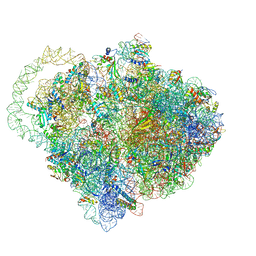 | |
5KK0
 
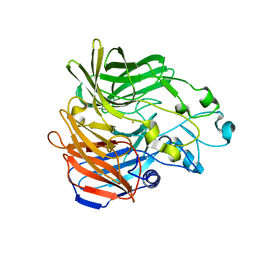 | | Synechocystis ACO mutant - T136A | | Descriptor: | Apocarotenoid-15,15'-oxygenase, CHLORIDE ION, FE (II) ION | | Authors: | Sui, X, Kiser, P.D, Palczewski, K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Utilization of Dioxygen by Carotenoid Cleavage Oxygenases.
J.Biol.Chem., 290, 2015
|
|
6W3W
 
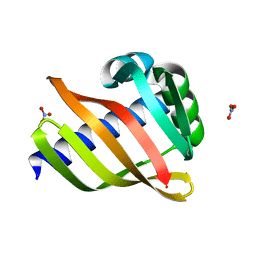 | | An enumerative algorithm for de novo design of proteins with diverse pocket structures | | Descriptor: | DENOVO NTF2, NITRATE ION | | Authors: | Bera, A.K, Basanta, B, Dimaio, F, Sankaran, B, Baker, D. | | Deposit date: | 2020-03-09 | | Release date: | 2020-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An enumerative algorithm for de novo design of proteins with diverse pocket structures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6NEJ
 
 | | scoulerine 9-O-methyltransferase from Thalictrum flavum complexed (13aS)-3,10-dimethoxy-5,8,13,13a-tetrahydro-6H-isoquino[3,2-a]isoquinoline-2,9-diol and with S-ADENOSYL-L-HOMOCYSTEINE | | Descriptor: | (13aS)-3,10-dimethoxy-5,8,13,13a-tetrahydro-6H-isoquino[3,2-a]isoquinoline-2,9-diol, (S)-scoulerine 9-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Valentic, T.R, Smolke, C.D, Payne, J.T. | | Deposit date: | 2018-12-17 | | Release date: | 2019-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional characterization of the scoulerine 9-O methyltransferase from Thalictrum flavum.
To Be Published
|
|
6YWP
 
 | | Structure of apo-CutA | | Descriptor: | CutA, MAGNESIUM ION | | Authors: | Malik, D, Kobylecki, K, Krawczyk, P, Poznanski, J, Jakielaszek, A, Napiorkowska, A, Dziembowski, A, Tomecki, R, Nowotny, M. | | Deposit date: | 2020-04-29 | | Release date: | 2020-08-05 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and mechanism of CutA, RNA nucleotidyl transferase with an unusual preference for cytosine.
Nucleic Acids Res., 48, 2020
|
|
5KX1
 
 | |
7WJU
 
 | | Cryo-EM structure of the AsCas12f1-sgRNAv1-dsDNA ternary complex | | Descriptor: | Non-target strand, OrfB_Zn_ribbon domain-containing protein, Target strand, ... | | Authors: | Wu, Z, Liu, D, Shen, H, Ji, Q. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure-directed functional evolution of the miniature CRISPR-AsCas12f1 system
To Be Published
|
|
4YT0
 
 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with 2-methyl-N-[3-(1-methylethoxy)phenyl]benzamide. | | Descriptor: | 2-methyl-N-[3-(1-methylethoxy)phenyl]benzamide, Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.66 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
8RSB
 
 | | p97 (VCP) mutant - F539A ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Arie, M, Matzov, D, Karmona, R, Szenkier, N, Stanhill, A, Navon, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | p97 (VCP) mutant - F539A ADP state
To Be Published
|
|
8FWI
 
 | | Structure of dodecameric KaiC-RS-S413E/S414E solved by cryo-EM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein KaiC, ... | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2023-01-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
8FWJ
 
 | | Structure of dodecameric KaiC-RS-S413E/S414E complexed with KaiB-RS solved by cryo-EM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein KaiB, ... | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2023-01-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
5KMD
 
 | | Structure of CavAb in complex with amlodipine | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein, ... | | Authors: | Tang, L, Gamal EL-Din, T.M, Swanson, T.M, Pryde, D.C, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2016-06-26 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs.
Nature, 537, 2016
|
|
4YO7
 
 | | Crystal Structure of an ABC transporter solute binding protein (IPR025997) from Bacillus halodurans C-125 (BH2323, TARGET EFI-511484) with bound myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-11 | | Release date: | 2015-03-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of an ABC transporter solute binding protein (IPR025997) from Bacillus halodurans C-125 (BH2323, TARGET EFI-511484) with bound myo-inositol
To be published
|
|
5L82
 
 | |
6CKG
 
 | | D-glycerate 3-kinase from Cryptococcus neoformans var. grubii serotype A (H99 / ATCC 208821 / CBS 10515 / FGSC 9487) | | Descriptor: | 1,2-ETHANEDIOL, D-glycerate 3-kinase | | Authors: | Horanyi, P.S, Abendroth, J, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-02-28 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D-glycerate 3-kinase from Cryptococcus neoformans var. grubii serotype A (H99 / ATCC 208821 / CBS 10515 / FGSC 9487)
To be Published
|
|
8IFG
 
 | | Cryo-EM structure of the Clr6S (Clr6-HDAC) complex from S. pombe | | Descriptor: | Chromatin modification-related protein eaf3, Cph1, Cph2, ... | | Authors: | Zhang, H.Q, Wang, X, Wang, Y.N, Liu, S.M, Zhang, Y, Xu, K, Ji, L.T, Kornberg, R.D. | | Deposit date: | 2023-02-17 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Class I histone deacetylase complex: Structure and functional correlates.
Proc Natl Acad Sci U S A, 120, 2023
|
|
6T14
 
 | | Native structure of mosquitocidal Cyt1A protoxin obtained by Serial Femtosecond Crystallography on in vivo grown crystals at pH 7 | | Descriptor: | SODIUM ION, Type-1Aa cytolytic delta-endotoxin | | Authors: | Tetreau, G, Banneville, A.S, Andreeva, E, Brewster, A.S, Hunter, M.S, Sierra, R.G, Young, I.D, Boutet, S, Coquelle, N, Cascio, D, Sawaya, M.R, Sauter, N.K, Colletier, J.P. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Serial femtosecond crystallography on in vivo-grown crystals drives elucidation of mosquitocidal Cyt1Aa bioactivation cascade.
Nat Commun, 11, 2020
|
|
8FON
 
 | | Crystal structure of tRNA^Lys(SUU) bound to AUA codon in the ribosomal P site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Nguyen, H.A, Hoffer, E.D, Maehigashi, T, Fagan, C.E, Dunham, C.M. | | Deposit date: | 2023-01-02 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structural basis for reduced ribosomal A-site fidelity in response to P-site codon-anticodon mismatches.
J.Biol.Chem., 299, 2023
|
|
5KRL
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the A-CD ring estrogen, (1S,7aS)-5-(2-chloro-4-hydroxyphenyl)-7a-methyl-2,3,3a,4,7,7a-hexahydro-1H-inden-1-ol | | Descriptor: | (1~{S},3~{a}~{R},7~{a}~{S})-5-(2-chloranyl-4-oxidanyl-phenyl)-2,3,3~{a},4,7,7~{a}-hexahydro-1~{H}-inden-1-ol, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
