7PJ9
 
 | |
6MAQ
 
 | | F9 Pilus Adhesin FmlH Lectin Domain from E. coli UTI89 in Complex with Galactoside 2'-{[(2S,3R,4R,5R,6R)-3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}-5-nitro-[1,1'-biphenyl]-3-carboxylic acid | | Descriptor: | 2'-{[2-(acetylamino)-2-deoxy-beta-D-galactopyranosyl]oxy}-5-nitro[1,1'-biphenyl]-3-carboxylic acid, Fimbrial adhesin FmlD, GLYCEROL | | Authors: | Klein, R.D, Hultgren, S.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Biphenyl Gal and GalNAc FmlH Lectin Antagonists of Uropathogenic E. coli (UPEC): Optimization through Iterative Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
6XNZ
 
 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Target Capture Complex | | Descriptor: | 12RSS integration strand (34-mer), 12RSS non-integration strand (34-mer), 23RSS integration strand (45-mer), ... | | Authors: | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
8OJM
 
 | | Galectin-3 in complex with 2,6-anhydro-3-deoxy-3-S-(beta-D-galactopyranosyl)-3-thio-D-glycero-D-galacto-heptonamide | | Descriptor: | (2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxane-2-carboxamide, 1-thio-beta-D-galactopyranose, Galectin-3, ... | | Authors: | Tsagkarakou, A.S, Leonidas, D.D. | | Deposit date: | 2023-03-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Strong Binding of C -Glycosylic1,2-Thiodisaccharides to Galectin-3─Enthalpy-Driven Affinity Enhancement by Water-Mediated Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
6M9W
 
 | | Structure of Mg-free KRAS4b (2-169) bound to GDP with the switch-I in fully open conformation | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Dharmaiah, S, Tran, T.H, Yan, W, Simanshu, D.K. | | Deposit date: | 2018-08-24 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of N-terminally processed KRAS provide insight into the role of N-acetylation.
Sci Rep, 9, 2019
|
|
6XNS
 
 | | C3_crown-05 | | Descriptor: | C3_crown-05 | | Authors: | Bick, M.J, Hsia, Y, Sankaran, B, Baker, D. | | Deposit date: | 2020-07-04 | | Release date: | 2020-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
8OJI
 
 | | Galectin-3 in complex with Methyl 2,6-anhydro-3-deoxy-3-S-(b-D-galactopyranosyl)-3-thio-D-glycero-L-altro-heptonate | | Descriptor: | 1-thio-beta-D-galactopyranose, CHLORIDE ION, Galectin-3, ... | | Authors: | Tsagkarakou, A.S, Leonidas, D.D. | | Deposit date: | 2023-03-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Strong Binding of C -Glycosylic1,2-Thiodisaccharides to Galectin-3─Enthalpy-Driven Affinity Enhancement by Water-Mediated Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
8OJK
 
 | | Galectin-3 in complex with 2,6-anhydro-3-deoxy-3-S-(beta-D-galactopyranosyl)-3-thio-D-glycero-L-altro-heptonamide | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxane-2-carboxamide, 1-thio-beta-D-galactopyranose, Galectin-3, ... | | Authors: | Tsagkarakou, A.S, Leonidas, D.D. | | Deposit date: | 2023-03-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Strong Binding of C -Glycosylic1,2-Thiodisaccharides to Galectin-3─Enthalpy-Driven Affinity Enhancement by Water-Mediated Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
8BTP
 
 | | Helical structure of BcThsA in complex with 1''-3'gc(etheno)ADPR | | Descriptor: | (1S,3S,4R,5R,7R,15R,16S,17R)-5-imidazo[2,1-f]purin-3-yl-10,12-bis(oxidanyl)-10,12-bis(oxidanylidene)-2,6,9,11,13,18-hexaoxa-10$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{3,7}]octadecane-4,16,17-triol, ETHENO-NAD, NAD(+) hydrolase ThsA | | Authors: | Tamulaitiene, G, Sasnauskas, G, Sabonis, D. | | Deposit date: | 2022-11-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Activation of Thoeris antiviral system via SIR2 effector filament assembly.
Nature, 627, 2024
|
|
8E04
 
 | | Structure of monomeric LRRK1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 1 | | Authors: | Reimer, J.M, Mathea, S, Chatterjee, D, Knapp, S, Leschziner, A.E. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of LRRK1 and mechanisms of autoinhibition and activation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5II8
 
 | | Orthorhombic crystal structure of red abalone lysin at 0.99 A resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Egg-lysin, SULFATE ION | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
6ME5
 
 | | XFEL crystal structure of human melatonin receptor MT1 in complex with agomelatine | | Descriptor: | OLEIC ACID, chimera protein of Melatonin receptor type 1A and GlgA glycogen synthase, ~{N}-[2-(7-methoxynaphthalen-1-yl)ethyl]ethanamide | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
6UCF
 
 | |
6O4L
 
 | | Structure of ALDH7A1 mutant E399D complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
7Y63
 
 | | ApoSIDT2-pH7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 2, ... | | Authors: | Gong, D.S. | | Deposit date: | 2022-06-18 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural insight into the human SID1 transmembrane family member 2 reveals its lipid hydrolytic activity.
Nat Commun, 14, 2023
|
|
7Y68
 
 | | SIDT2-pH5.5 plus miRNA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 2, ... | | Authors: | Gong, D.S. | | Deposit date: | 2022-06-18 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural insight into the human SID1 transmembrane family member 2 reveals its lipid hydrolytic activity.
Nat Commun, 14, 2023
|
|
7Y69
 
 | | ApoSIDT2-pH5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 2, ... | | Authors: | Gong, D.S. | | Deposit date: | 2022-06-18 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural insight into the human SID1 transmembrane family member 2 reveals its lipid hydrolytic activity.
Nat Commun, 14, 2023
|
|
6OTW
 
 | | Crystallographic Structure of (HbII-HbIII)-O2 from Lucina pectinata at pH 5.0 | | Descriptor: | Hemoglobin II, Hemoglobin III, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Marchany-Rivera, D, Smith, C.A, Rodriguez-Perez, J.D, Lopez-Garriga, J. | | Deposit date: | 2019-05-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Lucina pectinata oxyhemoglobin (II-III) heterodimer pH susceptibility.
J.Inorg.Biochem., 207, 2020
|
|
6OTY
 
 | | Crystallographic Structure of (HbII-HbIII)-O2 from Lucina pectinata at pH 4.0 | | Descriptor: | Hemoglobin II, Hemoglobin III, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Marchany-Rivera, D, Smith, C.A, Rodriguez-Perez, J.D, Lopez-Garriga, J. | | Deposit date: | 2019-05-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Lucina pectinata oxyhemoglobin (II-III) heterodimer pH susceptibility.
J.Inorg.Biochem., 207, 2020
|
|
5LFN
 
 | | Crystal structure of human chondroadherin | | Descriptor: | CHLORIDE ION, Chondroadherin | | Authors: | Ramisch, S, Pramhed, A, Logan, D.T. | | Deposit date: | 2016-07-03 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human chondroadherin: solving a difficult molecular-replacement problem using de novo models.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5IWI
 
 | | 1.98A structure of GSK945237 with S.aureus DNA gyrase and singly nicked DNA | | Descriptor: | (1R)-1-[(4-{[(6,7-dihydro[1,4]dioxino[2,3-c]pyridazin-3-yl)methyl]amino}piperidin-1-yl)methyl]-9-fluoro-1,2-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-4-one, CHLORIDE ION, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*AP*CP*CP*GP*CP*AP*CP*A)-3'), ... | | Authors: | Bax, B.D, Miles, T.J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel tricyclics (e.g., GSK945237) as potent inhibitors of bacterial type IIA topoisomerases.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8CHW
 
 | |
8CHT
 
 | |
6XNY
 
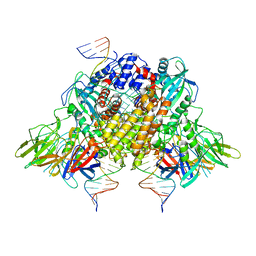 | | Structure of RAG1 (R848M/E649V)-RAG2-DNA Strand Transfer Complex (Paired-Form) | | Descriptor: | 12RSS integration strand (55-mer), 12RSS signal DNA top strand (34-mer), 23RSS integration strand (66-mer), ... | | Authors: | Zhang, Y, Corbett, E, Wu, S, Schatz, D.G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase.
Embo J., 39, 2020
|
|
8CHV
 
 | |
