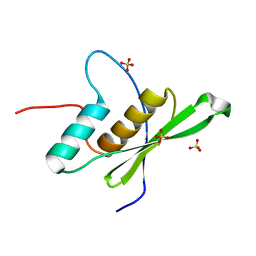
6EI8
 
 | |
6A8N
 
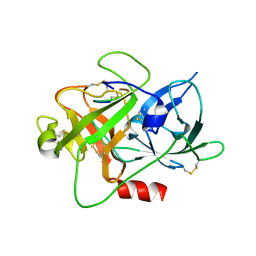 | | The crystal structure of muPAin-1-IG-2 in complex with muPA-SPD at pH8.5 | | Descriptor: | CYS-PRO-ALA-TYR-SER-ARG-TYR-ILE-GLY-CYS, Urokinase-type plasminogen activator B | | Authors: | Wang, D, Yang, Y.S, Jiang, L.G, Huang, M.D, Li, J.Y, Andreasen, P.A, Xu, P, Chen, Z. | | Deposit date: | 2018-07-09 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Suppression of Tumor Growth and Metastases by Targeted Intervention in Urokinase Activity with Cyclic Peptides.
J.Med.Chem., 62, 2019
|
|
3EPD
 
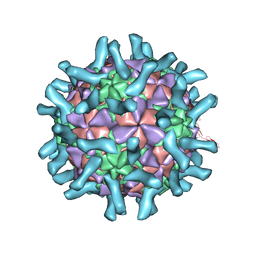 | | CryoEM structure of poliovirus receptor bound to poliovirus type 3 | | Descriptor: | MYRISTIC ACID, Poliovirus Type3 peptide, Poliovirus receptor, ... | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EPC
 
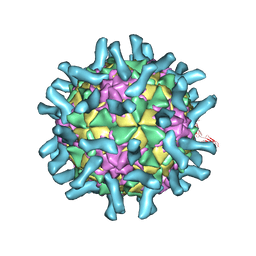 | | CryoEM structure of poliovirus receptor bound to poliovirus type 1 | | Descriptor: | MYRISTIC ACID, Poliovirus receptor, Protein VP1, ... | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EPF
 
 | | CryoEM structure of poliovirus receptor bound to poliovirus type 2 | | Descriptor: | 1[2-CHLORO-4-METHOXY-PHENYL-OXYMETHYL]-4-[2,6-DICHLORO-PHENYL-OXYMETHYL]-BENZENE, MYRISTIC ACID, Poliovirus receptor, ... | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7MF7
 
 | | Crystal structure of antibody 10E8v4-P100gA Fab | | Descriptor: | Antibody 10E8v4 Fab heavy chain, Antibody 10E8v4 Fab light chain | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2021-04-08 | | Release date: | 2021-07-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of HIV-1 Neutralizing Antibody 10E8 Delineate the Mechanistic Basis of Its Multi-Peak Behavior on Size-Exclusion Chromatography.
Antibodies, 10, 2021
|
|
5LU2
 
 | | Human 14-3-3 sigma complexed with long HSPB6 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, Heat shock protein beta-6 | | Authors: | Sluchanko, N.N, Beelen, S, Kulikova, A.A, Weeks, S.D, Antson, A.A, Gusev, N.B, Strelkov, S.V. | | Deposit date: | 2016-09-07 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Interaction of a Human Small Heat Shock Protein with the 14-3-3 Universal Signaling Regulator.
Structure, 25, 2017
|
|
8ADW
 
 | | Lipidic alpha-synuclein fibril - polymorph L1C | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-07-11 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
1CZH
 
 | | COMPARISONS OF WILD TYPE AND MUTANT FLAVODOXINS FROM ANACYSTIS NIDULANS. STRUCTURAL DETERMINANTS OF THE REDOX POTENTIALS. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, GLYCEROL, ... | | Authors: | Hoover, D.M, Drennan, C.L, Metzger, A.L, Osborne, C, Weber, C.H, Pattridge, K.A, Ludwig, M.L. | | Deposit date: | 1999-09-03 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials.
J.Mol.Biol., 294, 1999
|
|
7TRK
 
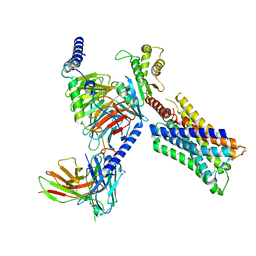 | | Human M4 muscarinic acetylcholine receptor complex with Gi1 and the agonist iperoxo | | Descriptor: | 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Antibody fragment scFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Vuckovic, Z, Mobbs, J.I, Belousoff, M.J, Glukhova, A, Sexton, P.M, Danev, R, Thal, D.M. | | Deposit date: | 2022-01-29 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and dynamic mechanisms of allostery at the M4 muscarinic acetylcholine receptor
Elife, 2023
|
|
7TRP
 
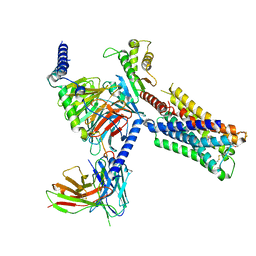 | | Human M4 muscarinic acetylcholine receptor complex with Gi1 and the agonist iperoxo and positive allosteric modulator LY2033298 | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-6-methoxy-4-methylthieno[2,3-b]pyridine-2-carboxamide, 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Antibody fragment scFv16, ... | | Authors: | Vuckovic, Z, Mobbs, J.I, Belousoff, M.J, Glukhova, A, Sexton, P.M, Danev, R, Thal, D.M. | | Deposit date: | 2022-01-30 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural and dynamic mechanisms of allostery at the M4 muscarinic acetylcholine receptor
Elife, 2023
|
|
3IVY
 
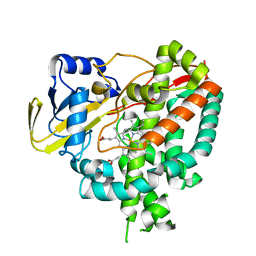 | | Crystal structure of Mycobacterium tuberculosis cytochrome P450 CYP125, p212121 crystal form | | Descriptor: | Cytochrome P450 CYP125, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | McLean, K.J, Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | The Structure of Mycobacterium tuberculosis CYP125: molecular basis for cholesterol binding in a P450 needed for host infection.
J.Biol.Chem., 284, 2009
|
|
1CRG
 
 | |
7TRQ
 
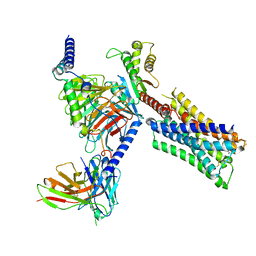 | | Human M4 muscarinic acetylcholine receptor complex with Gi1 and the agonist iperoxo and positive allosteric modulator VU0467154 | | Descriptor: | 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, 5-amino-3,4-dimethyl-N-{[4-(trifluoromethanesulfonyl)phenyl]methyl}thieno[2,3-c]pyridazine-6-carboxamide, Antibody fragment scFv16, ... | | Authors: | Vuckovic, Z, Mobbs, J.I, Belousoff, M.J, Glukhova, A, Sexton, P.M, Danev, R, Thal, D.M. | | Deposit date: | 2022-01-30 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural and dynamic mechanisms of allostery at the M4 muscarinic acetylcholine receptor
Elife, 2023
|
|
7TRS
 
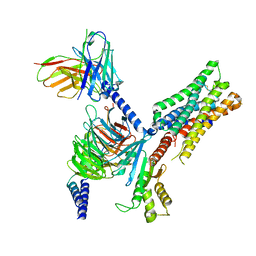 | | Human M4 muscarinic acetylcholine receptor complex with Gi1 and the endogenous agonist acetylcholine | | Descriptor: | ACETYLCHOLINE, Antibody fragment scFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Vuckovic, Z, Mobbs, J.I, Belousoff, M.J, Glukhova, A, Sexton, P.M, Danev, R, Thal, D.M. | | Deposit date: | 2022-01-30 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and dynamic mechanisms of allostery at the M4 muscarinic acetylcholine receptor
Elife, 2023
|
|
6RVV
 
 | | Structure of left-handed protein cage consisting of 24 eleven-membered ring proteins held together by gold (I) bridges. | | Descriptor: | GOLD ION, Transcription attenuation protein MtrB | | Authors: | Malay, A.D, Miyazaki, N, Biela, A.P, Iwasaki, K, Heddle, J.G. | | Deposit date: | 2019-06-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ultra-stable gold-coordinated protein cage displaying reversible assembly.
Nature, 569, 2019
|
|
8WP1
 
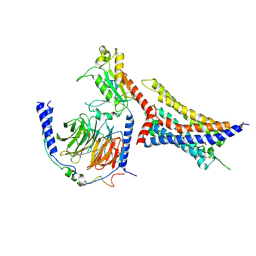 | | Cryo-EM structure of SUCR1 in complex with cis-epoxysuccinic acid and Gi proteins | | Descriptor: | (2R,3S)-oxirane-2,3-dicarboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, A, Ye, R.D. | | Deposit date: | 2023-10-08 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural insights into ligand recognition and activation of the succinate receptor SUCNR1.
Cell Rep, 43, 2024
|
|
3EXT
 
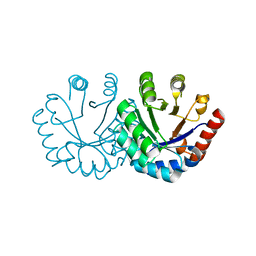 | | Crystal structure of KGPDC from Streptococcus mutans | | Descriptor: | MAGNESIUM ION, RmpD (Hexulose-6-phosphate synthase) | | Authors: | Liu, X, Li, G.L, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
6AIN
 
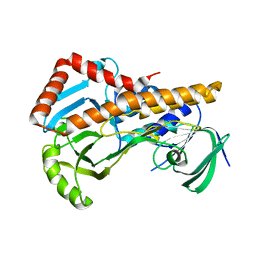 | | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PnpA | | Authors: | Chen, Q.Z, Huang, Y, Duan, Y.J, Li, Z.K, Liu, W.D, Cui, Z.L. | | Deposit date: | 2018-08-24 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of p-nitrophenol 4-monooxygenase PnpA from Pseudomonas putida DLL-E4: The key enzyme involved in p-nitrophenol degradation.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
4I5H
 
 | |
5XW2
 
 | |
1CZO
 
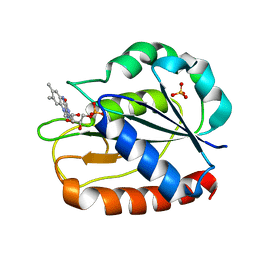 | | COMPARISONS OF WILD TYPE AND MUTANT FLAVODOXINS FROM ANACYSTIS NIDULANS. STRUCTURAL DETERMINANTS OF THE REDOX POTENTIALS. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, SULFATE ION | | Authors: | Hoover, D.M, Drennan, C.L, Metzger, A.L, Osborne, C, Weber, C.H, Pattridge, K.A, Ludwig, M.L. | | Deposit date: | 1999-09-03 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials.
J.Mol.Biol., 294, 1999
|
|
6AK5
 
 | | Binary Complex of Human DNA Polymerase Mu with MnGTP | | Descriptor: | DNA-directed DNA/RNA polymerase mu, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Chang, Y.K, Wu, W.J, Tsai, M.D. | | Deposit date: | 2018-08-30 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Human DNA Polymerase mu Can Use a Noncanonical Mechanism for Multiple Mn2+-Mediated Functions.
J.Am.Chem.Soc., 141, 2019
|
|
5G15
 
 | | Structure Aurora A (122-403) bound to activating monobody Mb1 and AMPPCP | | Descriptor: | AURORA A KINASE, MAGNESIUM ION, MB1 MONOBODY, ... | | Authors: | Zorba, A, Kutter, S, Kern, D, Koide, S, Koide, A. | | Deposit date: | 2016-03-23 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Allosteric modulation of a human protein kinase with monobodies.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8WVG
 
 | | Transporter bound with inhibitor | | Descriptor: | (3~{S},11~{b}~{R})-9,10-dimethoxy-3-(2-methylpropyl)-1,3,4,6,7,11~{b}-hexahydrobenzo[a]quinolizin-2-one, FabH, FabL, ... | | Authors: | Im, D, Iwata, S. | | Deposit date: | 2023-10-23 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Neurotransmitter recognition by human vesicular monoamine transporter 2.
Nat Commun, 15, 2024
|
|
