8SY8
 
 | | Crystal structure of TsaC | | Descriptor: | 4-formylbenzenesulfonate dehydrogenase TsaC | | Authors: | Boggs, D.G, Tian, J, Bridwell-Rabb, J. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The NADH recycling enzymes TsaC and TsaD regenerate reducing equivalents for Rieske oxygenase chemistry.
J.Biol.Chem., 299, 2023
|
|
6XUZ
 
 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 4 | | Descriptor: | 6-[1-[(2~{S})-1-methoxypropan-2-yl]-6-[(3~{S})-3-methylmorpholin-4-yl]imidazo[4,5-c]pyridin-2-yl]-3-methyl-~{N}-propan-2-yl-[1,2,4]triazolo[4,3-a]pyrazin-8-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7URS
 
 | |
8X72
 
 | | The Crystal Structure of PLK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Wu, B. | | Deposit date: | 2023-11-22 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of PLK1 from Biortus.
To Be Published
|
|
6VRD
 
 | | Crystal structure of RNase H/RNA/PS-ASO complex at an atomic level | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, RNA (5'-R(*(OMC)P*(N7X)P*(T39)P*(C5L)P*(A2M))-D(P*(SC)P*(PST)P*(SC)P*(AS)P*(SC)P*(SC)P*(SC)P*(AS)P*(SC)P*(PST))-R(P*(6OO)P*(RFJ)P*(6OO)P*(6OO)P*(6NW))-3'), ... | | Authors: | Cho, Y.-J, Butler, D. | | Deposit date: | 2020-02-07 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Crystal structure of RNase H/RNA/PS-ASO complex at an atomic level
To Be Published
|
|
8X23
 
 | | The Crystal Structure of MAPK13 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Mitogen-activated protein kinase 13 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Pan, W. | | Deposit date: | 2023-11-09 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of MAPK13 from Biortus.
To Be Published
|
|
7URQ
 
 | |
6TP4
 
 | | Crystal structure of the Orexin-1 receptor in complex with ACT-462206 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2~{S})-~{N}-(3,5-dimethylphenyl)-1-(4-methoxyphenyl)sulfonyl-pyrrolidine-2-carboxamide, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6CDM
 
 | |
8X2A
 
 | | The Crystal Structure of BMX from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3S)-3-{[(2E)-but-2-enoyl]amino}piperidin-1-yl]-5-fluoro-2,3-dimethyl-1H-indole-7-carboxamide, CHLORIDE ION, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Pan, W. | | Deposit date: | 2023-11-09 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Crystal Structure of BMX from Biortus.
To Be Published
|
|
8P32
 
 | | BB0238 from Borrelia burgdorferi, Se-Met data for Leu240Met mutant | | Descriptor: | BB0238 | | Authors: | Brangulis, K, Foor, S.D, Shakya, A.K, Rana, V.S, Bista, S, Kitsou, C, Ronzetti, M, Linden, S.B, Altieri, A.S, Akopjana, I, Baljinnyam, B, Nelson, D.C, Simeonov, A, Herzberg, O, Caimano, M.J, Pal, U. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A unique borrelial protein facilitates microbial immune evasion.
Mbio, 14, 2023
|
|
8TAU
 
 | | APC/C-CDH1-UBE2C-UBE2S-Ubiquitin-CyclinB | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Bodrug, T, Welsh, K.A, Bolhuis, D.L, Paulakonis, E, Martinez-Chacin, R.C, Liu, B, Pinkin, N, Bonacci, T, Cui, L, Xu, P, Roscow, O, Amann, S.J, Grishkovskaya, I, Emanuele, M.J, Harrison, J.S, Steimel, J.P, Hahn, K.M, Zhang, W, Zhong, E, Haselbach, D, Brown, N.G. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C).
Nat.Struct.Mol.Biol., 30, 2023
|
|
6Q41
 
 | | Atomic resolution crystal structure of a BAA collagen heterotrimer | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Leading chain of the BAA collagen heterotrimer, ... | | Authors: | Jalan, A.A, Hartgerink, J.D, Brear, P, Leitinger, B, Farndale, R.W. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Atomic resolution crystal structure of an AAB collagen heterotrimer
Nat.Chem.Biol., 2019
|
|
6VYK
 
 | | Cryo-EM structure of mechanosensitive channel MscS in PC-18:1 nanodiscs | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mechanosensitive channel MscS | | Authors: | Zhang, Y, Daday, C, Gu, R, Cox, C.D, Martinac, B, Groot, B, Walz, T. | | Deposit date: | 2020-02-27 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualization of the mechanosensitive ion channel MscS under membrane tension.
Nature, 590, 2021
|
|
7ND2
 
 | | Cryo-EM structure of the human FERRY complex | | Descriptor: | Glutamine amidotransferase-like class 1 domain-containing protein 1, Protein phosphatase 1 regulatory subunit 21, Quinone oxidoreductase-like protein 1 | | Authors: | Quentin, D, Klink, B.U, Raunser, S. | | Deposit date: | 2021-01-29 | | Release date: | 2022-03-02 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of mRNA binding by the human FERRY Rab5 effector complex.
Mol.Cell, 83, 2023
|
|
6VPZ
 
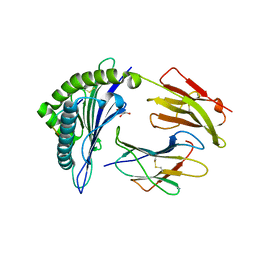 | | HLA-B*27:05 presenting an HIV-1 11mer peptide | | Descriptor: | 11-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6IBW
 
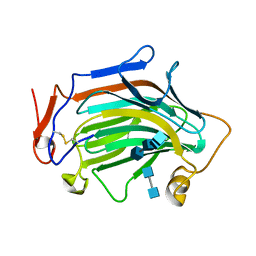 | | Crh5 transglycosylase in complex with NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Probable glycosidase crf1 | | Authors: | Fang, W, Bartual, S.G, van Aalten, D.M.F. | | Deposit date: | 2018-12-01 | | Release date: | 2019-02-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanisms of redundancy and specificity of the Aspergillus fumigatus Crh transglycosylases.
Nat Commun, 10, 2019
|
|
7JUO
 
 | | CBP bromodomain complexed with YF2-23 | | Descriptor: | CREB-binding protein, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(1-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-3-yl)-1H-indol-4-yl}ethanesulfonamide | | Authors: | Ratia, K.M, Xiong, R, Principe, D, Li, Y, Huang, F, Rana, A, Thatcher, G. | | Deposit date: | 2020-08-20 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | XP-524 is a dual-BET/EP300 inhibitor that represses oncogenic KRAS and potentiates immune checkpoint inhibition in pancreatic cancer.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3MHV
 
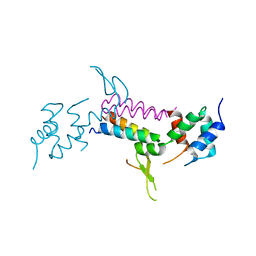 | | Crystal Structure of Vps4 and Vta1 | | Descriptor: | Vacuolar protein sorting-associated protein 4, Vacuolar protein sorting-associated protein VTA1 | | Authors: | Yang, D, Hurley, J.H. | | Deposit date: | 2010-04-09 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural role of the Vps4-Vta1 interface in ESCRT-III recycling
Structure, 18, 2010
|
|
6XCH
 
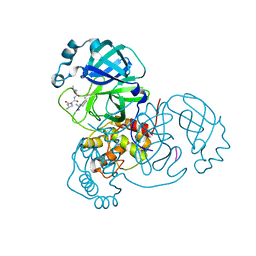 | |
8OU3
 
 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 8d | | Descriptor: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2-fluoranyl-benzamide, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Heim, C, Bischof, L, Maiwald, S, Hartmann, M.D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
8T1Z
 
 | |
8X5L
 
 | | The Crystal Structure of PRKACA from Biortus. | | Descriptor: | (2S)-2-(4-chlorophenyl)-2-hydroxy-2-[4-(1H-pyrazol-4-yl)phenyl]ethanaminium, SODIUM ION, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Wang, F, Cheng, W, Lv, Z, Lin, D, Pan, W. | | Deposit date: | 2023-11-17 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of PRKACA from Biortus.
To Be Published
|
|
6XWQ
 
 | |
8T24
 
 | |
