8Q3C
 
 | | Structure of Selenomonas ruminantium lactate dehydrogenase I85R mutant | | Descriptor: | CHLORIDE ION, L-lactate dehydrogenase, NITRATE ION, ... | | Authors: | Bertrand, Q, Coquille, S, Iorio, A, Sterpone, F, Madern, D. | | Deposit date: | 2023-08-03 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical, structural and dynamical characterizations of the lactate dehydrogenase from Selenomonas ruminantium provide information about an intermediate evolutionary step prior to complete allosteric regulation acquisition in the super family of lactate and malate dehydrogenases.
J.Struct.Biol., 215, 2023
|
|
8QFH
 
 | | Room temperature crystal structure of the Photoactivated Adenylate Cyclase OaPAC with ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
4EBW
 
 | | Structure of Focal Adhesion Kinase catalytic domain in complex with novel allosteric inhibitor | | Descriptor: | 1-ethyl-8-(4-ethylphenyl)-5-methyl-1,5-dihydropyrazolo[4,3-c][2,1]benzothiazine 4,4-dioxide, Focal adhesion kinase 1 | | Authors: | Iwatani, M, Iwata, H, Okabe, A, Skene, R.J, Tomita, N, Hayashi, Y, Aramaki, Y, Hosfield, D.J, Hori, A, Baba, A, Miki, H. | | Deposit date: | 2012-03-25 | | Release date: | 2012-07-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and characterization of novel allosteric FAK inhibitors.
Eur.J.Med.Chem., 61, 2013
|
|
8QFJ
 
 | | Room temperature crystal structure of the Photoactivated Adenylate Cyclase OaPAC after blue light excitation at 2.3 us delay | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
7NXU
 
 | | Crystal structure of the tick-borne encephalitis virus NS3 helicase in complex with ADP-Pi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, NS3 helicase domain, ... | | Authors: | Anindita, P.D, Grinkevich, P, Franta, Z. | | Deposit date: | 2021-03-19 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic insight into the RNA-stimulated ATPase activity of tick-borne encephalitis virus helicase.
J.Biol.Chem., 298, 2022
|
|
8QFE
 
 | | Cryogenic crystal structure of the Photoactivated Adenylate Cyclase OaPAC | | Descriptor: | FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, MAGNESIUM ION | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
8V1S
 
 | | Herpes simplex virus 1 polymerase holoenzyme bound to mismatched DNA in editing conformation | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
3KYR
 
 | | Bace-1 in complex with a norstatine type inhibitor | | Descriptor: | 3-[[(2S)-2-[[[(2S)-2-[[(2S)-2-[[(2S)-2-azanyl-3-(1H-1,2,3,4-tetrazol-5-ylcarbonylamino)propanoyl]amino]-3-methyl-butanoyl]amino]-4-methyl-pentanoyl]amino]methyl]-2-hydroxy-4-phenyl-butanoyl]amino]benzoic acid, Beta-secretase 1 | | Authors: | Lindberg, J.D, Borkakoti, N, Derbyshire, D, Nystrom, S. | | Deposit date: | 2009-12-07 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigation of a-phenylnorstatine and a-benzylnorstatine as transition state isostere motifs in the search for new BACE-1 inhibiotrs
To be Published
|
|
8QFI
 
 | | Room temperature crystal structure of the Photoactivated Adenylate Cyclase OaPAC after blue light excitation at 1.8 us delay | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Sato, T, Round, A, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
6ZPA
 
 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dA and one catalytic Zn2+ ion | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, DatZ, LITHIUM ION, ... | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.86000258 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
8BHU
 
 | | Crystal Structure of SilF (Ag(I) form | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SILVER ION, SULFATE ION, ... | | Authors: | Lithgo, R.M, Carr, S.B, Quigley, A.M, Scott, D.J. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of SilF (apo form)
To Be Published
|
|
8QFF
 
 | | Cryogenic crystal structure of the Photoactivated Adenylate Cyclase OaPAC with ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
6ZPC
 
 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DatZ, LITHIUM ION, ... | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.2683593 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
7NFO
 
 | |
8BIT
 
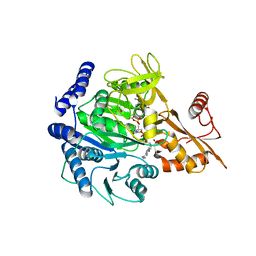 | | Crystal structure of acyl-CoA synthetase from Metallosphaera sedula in complex with Coenzyme A and acetyl-AMP | | Descriptor: | 4-hydroxybutyrate--CoA ligase 1, COENZYME A, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] ethanoate | | Authors: | Capra, N, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2022-11-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Adapting an acyl CoA ligase from Metallosphaera sedula for lactam formation by structure-guided protein engineering
Front Catal, 4, 2024
|
|
6ZP9
 
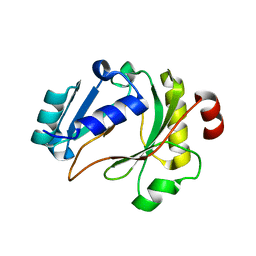 | |
6ZPB
 
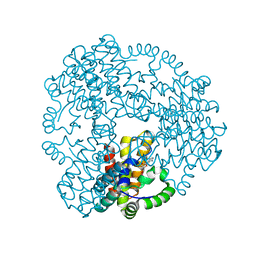 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dA and two catalytic Co2+ ions | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, COBALT (II) ION, DatZ | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.72097385 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
7NFF
 
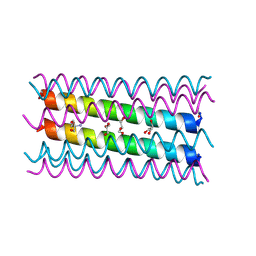 | | An octameric barrel state of a de novo coiled-coil assembly: CC-Type2-(LaId)4-I24A. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CC-Type2-(LaId)4-I24A, GLYCEROL | | Authors: | Rhys, G.G, Dawson, W.M, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2021-02-07 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Differential sensing with arrays of de novo designed peptide assemblies.
Nat Commun, 14, 2023
|
|
7NFL
 
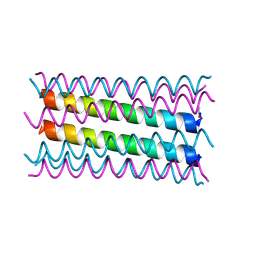 | |
6XO1
 
 | | Crystal structure of N97A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.758 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
8BJR
 
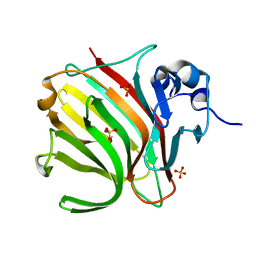 | |
8RS3
 
 | | Thaumatin measured via serial crystallography from a kapton HARE-chip (50 micron) | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Bosman, R, Prester, A, von Soosten, L, Dibenedetto, S, Bartels, K, Sung, S, von Stetten, D, Mehrabi, P, Schulz, E.C. | | Deposit date: | 2024-01-24 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kapton-based HARE chips for fixed-target serial crystallography
To Be Published
|
|
7NFK
 
 | | An octameric barrel state of a de novo coiled-coil assembly: CC-Type2-(LaId)4-I24S. | | Descriptor: | CC-Type2-(LaId)4-I24S, ISOPROPYL ALCOHOL | | Authors: | Rhys, G.G, Dawson, W.M, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2021-02-07 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Differential sensing with arrays of de novo designed peptide assemblies.
Nat Commun, 14, 2023
|
|
6IZV
 
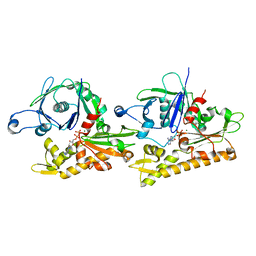 | | Averaged strand structure of a 15-stranded ParM filament from Clostridium botulinum | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative plasmid segregation protein ParM | | Authors: | Koh, F, Narita, A, Lee, L.J, Tan, Y.Z, Dandey, V.P, Tanaka, K, Popp, D, Robinson, R.C. | | Deposit date: | 2018-12-20 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structure of a 15-stranded actin-like filament from Clostridium botulinum.
Nat Commun, 10, 2019
|
|
6MUR
 
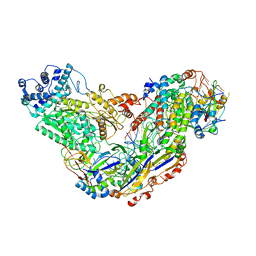 | | Cryo-EM structure of Csm-crRNA-target RNA ternary complex in type III-A CRISPR-Cas system | | Descriptor: | RNA (27-MER), RNA (5'-R(P*GP*CP*AP*AP*AP*CP*CP*GP*CP*CP*UP*CP*UP*GP*CP*CP*CP*GP*C)-3'), Uncharacterized protein, ... | | Authors: | Jia, N, Wang, C, Eng, E.T, Patel, D.J. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
