2L4S
 
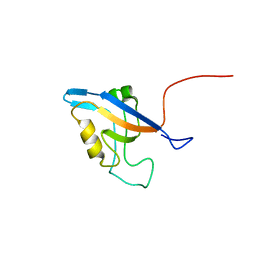 | | Promiscuous Binding at the Crossroads of Numerous Cancer Pathways: Insight from the Binding of GIP with Glutaminase L | | Descriptor: | Tax1-binding protein 3 | | Authors: | Zoetewey, D.L, Ovee, M, Banerjee, M, Bhaskaran, R, Mohanty, S. | | Deposit date: | 2010-10-13 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous binding at the crossroads of numerous cancer pathways: insight from the binding of glutaminase interacting protein with glutaminase L.
Biochemistry, 50, 2011
|
|
7JNO
 
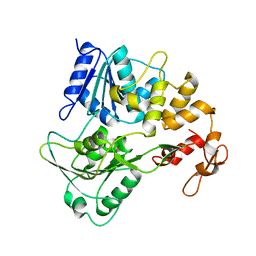 | |
2LRH
 
 | | Solution NMR structure of de novo designed protein, p-loop ntpase fold, Northeast Structural Genomics Consortium target or137 | | Descriptor: | De novo designed protein | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Lee, H, Janjua, H, Kohan, E, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-03-30 | | Release date: | 2012-07-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of de novo designed protein, p-loop ntpase fold, Northeast Structural Genomics Consortium target or137
To be Published
|
|
5DYG
 
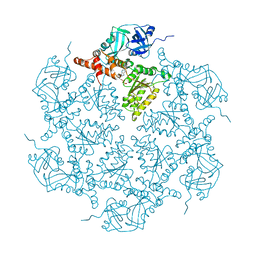 | | Structure of p97 N-D1 L198W mutant in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Tang, W.K, Xia, D. | | Deposit date: | 2015-09-24 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of the D1-D2 Linker of Human VCP/p97 in the Asymmetry and ATPase Activity of the D1-domain.
Sci Rep, 6, 2016
|
|
2LJ3
 
 | |
2L7N
 
 | | Solution Structure of the R5 Domain of Talin | | Descriptor: | Talin-1 | | Authors: | Goult, B.T, Gingras, A.R, Bate, N, Barsukov, I.L, Roberts, G.C.K, Critchley, D.R. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover.
J.Biol.Chem., 288, 2013
|
|
6E5J
 
 | | Heterogeneous-Backbone Mimics of a Designed Disulfide-Rich Protein: Aib turn, beta3 helix, N-methyl hairpin | | Descriptor: | Designed peptide NC_HEE_D1: Aib turn, beta3 helix, N-methyl hairpin mutant | | Authors: | Cabalteja, C.C, Mihalko, D.S, Horne, W.S. | | Deposit date: | 2018-07-20 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Foldamer Mimics of a Computationally Designed, Disulfide-Rich Miniprotein.
Chembiochem, 20, 2019
|
|
3G7C
 
 | | Structure of the Phosphorylation Mimetic of Occludin C-term Tail | | Descriptor: | Occludin | | Authors: | Tash, B.R, Sundstrom, J.M, Murakami, T, Flanagan, J.M, Bewley, M.C, Antonetii, D.A. | | Deposit date: | 2009-02-09 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and analysis of occludin phosphosites: a combined mass spectrometry and bioinformatics approach.
J.PROTEOME RES., 8, 2009
|
|
2L4T
 
 | | GIP/Glutaminase L peptide complex | | Descriptor: | Glutaminase L peptide, Tax1-binding protein 3 | | Authors: | Zoetewey, D.L, Ovee, M, Banerjee, M, Bhaskaran, R, Mohanty, S. | | Deposit date: | 2010-10-13 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Promiscuous binding at the crossroads of numerous cancer pathways: insight from the binding of glutaminase interacting protein with glutaminase L.
Biochemistry, 50, 2011
|
|
2FNB
 
 | | NMR STRUCTURE OF THE FIBRONECTIN ED-B DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (FIBRONECTIN) | | Authors: | Fattorusso, R, Pellecchia, M, Viti, F, Neri, P, Neri, D, Wuthrich, K. | | Deposit date: | 1998-12-16 | | Release date: | 1998-12-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the human oncofoetal fibronectin ED-B domain, a specific marker for angiogenesis.
Structure Fold.Des., 7, 1999
|
|
3FKG
 
 | | Crystal Structure Analysis of Fungal Versatile Peroxidase from Pleurotus eryngii | | Descriptor: | CACODYLATE ION, CALCIUM ION, FE (III) ION, ... | | Authors: | Piontek, K, Martinez, A.T, Choinowski, T, Plattner, D.A. | | Deposit date: | 2008-12-16 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Site-directed Mutagenesis Study of Versatile Peroxidase Oxidizing both Mn(II) and Aromatic Substrates
To be Published
|
|
2L5G
 
 | | Co-ordinates and 1H, 13C and 15N chemical shift assignments for the complex of GPS2 53-90 and SMRT 167-207 | | Descriptor: | G protein pathway suppressor 2, Putative uncharacterized protein NCOR2 | | Authors: | Oberoi, J, Yang, J, Neuhaus, D, Schwabe, J.W.R. | | Deposit date: | 2010-11-01 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2LLM
 
 | |
3HAG
 
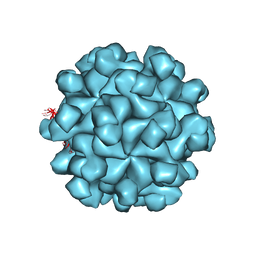 | | Crystal structure of the Hepatitis E Virus-like Particle | | Descriptor: | Capsid protein | | Authors: | Guu, T.S.Y, Liu, Z, Ye, Q, Mata, D.A, Li, K, Yin, C, Zhang, J, Tao, Y.J. | | Deposit date: | 2009-05-01 | | Release date: | 2009-09-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2LEW
 
 | | Structural Plasticity of Paneth cell alpha-Defensins: Characterization of Salt-Bridge Deficient Analogues of Mouse Cryptdin-4 | | Descriptor: | Alpha-defensin 4 | | Authors: | Rosengren, K, Andersson, H.S, Haugaard-Kedstrom, L.M, Bengtsson, E, Daly, N.L, Craik, D.J. | | Deposit date: | 2011-06-24 | | Release date: | 2012-05-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The alpha-defensin salt-bridge induces backbone stability to facilitate folding and confer proteolytic resistance.
Amino Acids, 43, 2012
|
|
7K3D
 
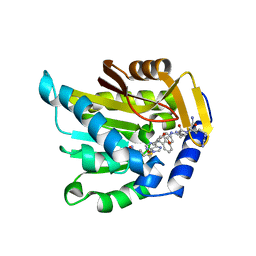 | | The structure of NTMT1 in complex with compound DC1-13 | | Descriptor: | N-terminal Xaa-Pro-Lys N-methyltransferase 1, N~2~-{(2S)-1-[(naphthalen-1-yl)acetyl]-2,5-dihydro-1H-pyrrole-2-carbonyl}-L-lysyl-L-argininamide, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Chen, D, Huang, R, Noinaj, N. | | Deposit date: | 2020-09-11 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure-based Discovery of Cell-Potent Peptidomimetic Inhibitors for Protein N-Terminal Methyltransferase 1.
Acs Med.Chem.Lett., 12, 2021
|
|
7NBA
 
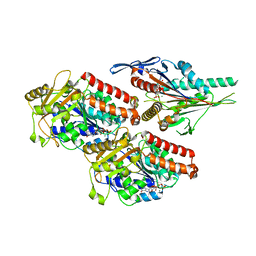 | | Plasmodium falciparum kinesin-5 motor domain bound to AMPPNP, complexed with 14 protofilament microtubule. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Kinesin motor domain-containing protein,Kinesin motor domain-containing protein, MAGNESIUM ION, ... | | Authors: | Cook, A.D, Roberts, A, Atherton, J, Tewari, R, Topf, M, Moores, C.A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of a microtubule-bound parasite kinesin motor and implications for its mechanism and inhibition.
J.Biol.Chem., 297, 2021
|
|
2LGN
 
 | | Lactococcin 972 | | Descriptor: | Lactococcin 972 | | Authors: | Turner, D.L, Lamosa, P, Martinez, B. | | Deposit date: | 2011-07-29 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and properties of lactococcin 972 from Lactococcus lactis
To be Published
|
|
2LPT
 
 | |
2LKR
 
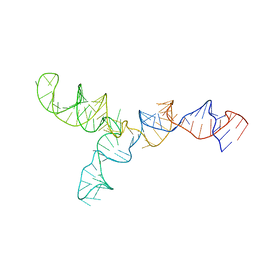 | | Yeast U2/U6 complex | | Descriptor: | RNA (111-MER) | | Authors: | Burke, J.E, Sashital, D.G, Zuo, X, Wang, Y, Butcher, S.E. | | Deposit date: | 2011-10-19 | | Release date: | 2012-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the yeast U2/U6 snRNA complex.
Rna, 18, 2012
|
|
8R3X
 
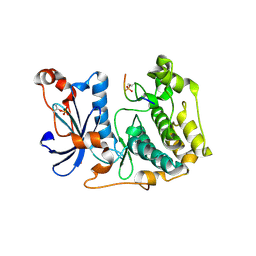 | | Crystal structure of aPKC Iota kinase domain with LLGL2 peptide | | Descriptor: | LLGL scribble cell polarity complex component 2, Protein kinase C iota type | | Authors: | Soriano, E.V, Earl, C.P, Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2023-11-10 | | Release date: | 2024-11-20 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Capture, mutual inhibition and release mechanism for aPKC-Par6 and its multisite polarity substrate Lgl.
Nat.Struct.Mol.Biol., 32, 2025
|
|
2LN3
 
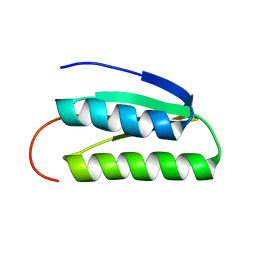 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, IF3-like fold, Northeast Structural Genomics Consortium Target OR135 (CASD target) | | Descriptor: | DE NOVO DESIGNED PROTEIN OR135 | | Authors: | Liu, G, Koga, R, Koga, N, Xiao, R, Lee, H, Janjua, H, Kohan, E, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-15 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
8UHI
 
 | | Structure of the far-red light-absorbing allophycocyanin core expressed during FaRLiP | | Descriptor: | MAGNESIUM ION, RIBULOSE-1,5-DIPHOSPHATE, Ribulose bisphosphate carboxylase large subunit, ... | | Authors: | Gisriel, C.J, Bryant, D.A, Brudvig, G.W, Shen, G. | | Deposit date: | 2023-10-09 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structure of the antenna complex expressed during far-red light photoacclimation in Synechococcus sp. PCC 7335.
J.Biol.Chem., 300, 2023
|
|
2LTA
 
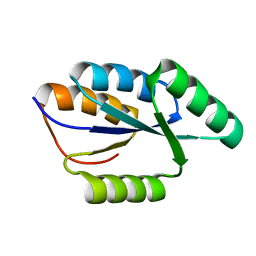 | | Solution NMR structure of De novo designed protein, rossmann 3x1 fold, Northeast Structural Genomics Consortium target OR157 | | Descriptor: | De novo designed protein | | Authors: | Liu, G, Koga, R, Koga, N, Xiao, R, Pederson, K, Hamilton, K, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
5FUA
 
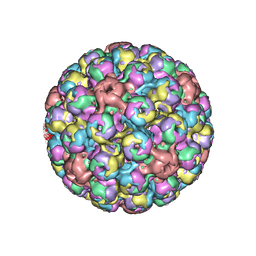 | | Cryo-EM of BK polyomavirus | | Descriptor: | MAJOR CAPSID PROTEIN VP1 | | Authors: | Hurdiss, D.L, Morgan, E.L, Thompson, R.F, Prescott, E.L, Panou, M.M, Macdonald, A, Ranson, N.A. | | Deposit date: | 2016-01-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | New Structural Insights Into the Genome and Minor Capsid Proteins of Bk Polyomavirus Using Cryo-Electron Microscopy.
Structure, 24, 2016
|
|
