6UVR
 
 | |
8OSF
 
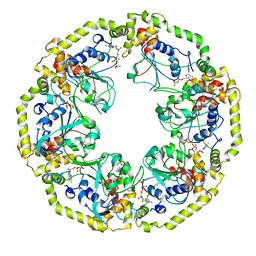 | | AAA+ motor subunit ChlI of magnesium chelatase, hexamer conformation A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shvarev, D, Moeller, A. | | Deposit date: | 2023-04-18 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational variability of cyanobacterial ChlI, the AAA+ motor of magnesium chelatase involved in chlorophyll biosynthesis.
Mbio, 14, 2023
|
|
8QUA
 
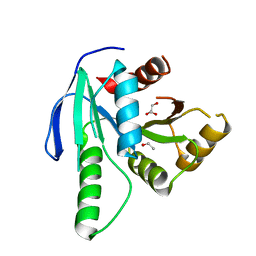 | | GTP binding protein YsxC from Staphylococcus aureus | | Descriptor: | ACETYL GROUP, GLYCEROL, Probable GTP-binding protein EngB | | Authors: | Biktimirov, A, Islamov, D, Lazarenko, V, Fatkhullin, B, Validov, S, Yusupov, M, Usachev, K. | | Deposit date: | 2023-10-15 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GTPase YsxC from Staphylococcus aureus.
Biochem.Biophys.Res.Commun., 699, 2024
|
|
5I3O
 
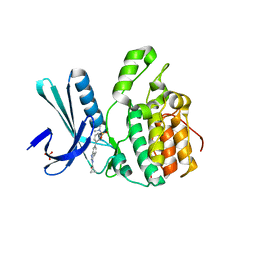 | | Crystal Structure of BMP-2-inducible kinase in complex with an Indazole inhibitor | | Descriptor: | BMP-2-inducible protein kinase, N-(6-{3-[(dimethylsulfamoyl)amino]phenyl}-1H-indazol-3-yl)cyclopropanecarboxamide, SULFATE ION | | Authors: | Counago, R.M, Sorrell, F.J, Krojer, T, Savitsky, P, Elkins, J.M, Axtman, A, Drewry, D, Wells, C, Zhang, C, Zuercher, W, Willson, T.M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Arruda, P, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of BMP-2-inducible kinase in complex with an Indazole inhibitor
To Be Published
|
|
5VFT
 
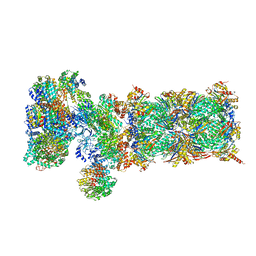 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
2LV7
 
 | | Solution structure of Ca2+-bound CaBP7 N-terminal doman | | Descriptor: | CALCIUM ION, Calcium-binding protein 7 | | Authors: | Mccue, H.V, Patel, P, Herbert, A.P, Lian, L, Burgoyne, R.D, Haynes, L.P. | | Deposit date: | 2012-06-29 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Ca2+-bound N-terminal Domain of CaBP7: A REGULATOR OF GOLGI TRAFFICKING.
J.Biol.Chem., 287, 2012
|
|
2LVR
 
 | | Solution structure of Miz-1 zinc finger 8 | | Descriptor: | ZINC ION, Zinc finger and BTB domain-containing protein 17 | | Authors: | Bedard, M, Maltais, L, Beaulieu, M, Bernard, D, Lavigne, P. | | Deposit date: | 2012-07-10 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure note: solution structure of human Miz-1 zinc fingers 8 to 10.
J.Biomol.Nmr, 54, 2012
|
|
6CLA
 
 | | 2.80 A MicroED structure of proteinase K at 6.0 e- / A^2 | | Descriptor: | Proteinase K | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
2LVB
 
 | | Solution NMR Structure DE NOVO DESIGNED PFK fold PROTEIN, Northeast Structural Genomics Consortium (NESG) Target OR250 | | Descriptor: | DE NOVO DESIGNED PFK fold PROTEIN | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Hamilton, K, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-30 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
2LVT
 
 | | Solution structure of Miz-1 zinc finger 9 | | Descriptor: | ZINC ION, Zinc finger and BTB domain-containing protein 17 | | Authors: | Bedard, M, Maltais, L, Beaulieu, M, Bernard, D, Lavigne, P. | | Deposit date: | 2012-07-11 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure note: solution structure of human Miz-1 zinc fingers 8 to 10.
J.Biomol.Nmr, 54, 2012
|
|
2LU3
 
 | |
2M0A
 
 | | Solution structure of MHV nsp3a | | Descriptor: | Non-structural protein 3 | | Authors: | Keane, S.C, Giedroc, D.P. | | Deposit date: | 2012-10-23 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mouse Hepatitis Virus (MHV) nsp3a and Determinants of the Interaction with MHV Nucleocapsid (N) Protein.
J.Virol., 87, 2013
|
|
6UVT
 
 | |
2LUR
 
 | |
2LW4
 
 | | Solution NMR Structure of Human Transcription Elongation Factor A protein 2, Central Domain, Northeast Structural Genomics Consortium (NESG) Target HR8682B | | Descriptor: | Transcription elongation factor A protein 2 | | Authors: | Eletsky, A, Wang, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-07-20 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Human Transcription Elongation Factor A protein 2, Central Domain (CASP Target)
To be Published
|
|
2MLD
 
 | |
2M0B
 
 | | Homodimeric transmembrane domain of the human receptor tyrosine kinase ErbB1 (EGFR, HER1) in micelles | | Descriptor: | Epidermal growth factor receptor | | Authors: | Lesovoy, D.M, Bocharov, E.V, Pustovalova, Y.E, Bocharova, O.V, Arseniev, A.S. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Alternative packing of EGFR transmembrane domain suggests that protein-lipid interactions underlie signal conduction across membrane.
Biochim. Biophys. Acta, 1858, 2016
|
|
8SKG
 
 | |
2M3U
 
 | |
5I3R
 
 | | Crystal Structure of BMP-2-inducible kinase in complex with an Indazole inhibitor | | Descriptor: | BMP-2-inducible protein kinase, N-[6-(3-{[(cyclopropylmethyl)sulfonyl]amino}phenyl)-1H-indazol-3-yl]cyclopropanecarboxamide, PHOSPHATE ION | | Authors: | Counago, R.M, Sorrell, F.J, Krojer, T, Savitsky, P, Elkins, J.M, Axtman, A, Drewry, D, Wells, C, Zhang, C, Zuercher, W, Willson, T.M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Arruda, P, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of BMP-2-inducible kinase in complex with an Indazole inhibitor
To Be Published
|
|
8C1E
 
 | | Aurora A kinase in complex with TPX2-inhibitor 9 | | Descriptor: | 4-(4-chloranyl-3-cyano-phenyl)-7-methyl-1~{H}-indole-6-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, ... | | Authors: | Fischer, G, Rocaboy, M, Blaszczyk, B, Moschetti, T, Wang, X, Scott, D.E, Coyne, A.G, Dagostin, C, Rooney, T, Bayly, A, Feng, J, Asteian, A, Alcaide-Lopez, A, Stockwell, S, Skidmore, J, Venkitaraman, A.R, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Selective Aurora A-TPX2 Interaction Inhibitors Have In Vivo Efficacy as Targeted Antimitotic Agents.
J.Med.Chem., 67, 2024
|
|
9EDP
 
 | | GVIII-Chiba040502 norovirus protruding domain | | Descriptor: | CHLORIDE ION, VP1 | | Authors: | Holroyd, D.L, Kumar, A, Bruning, J.B, Hansman, G.S. | | Deposit date: | 2024-11-17 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Antigenic structural analysis of bat and human norovirus protruding (P) domains.
J.Virol., 99, 2025
|
|
7AZP
 
 | |
2M5B
 
 | | The NMR structure of the BID-BAK complex | | Descriptor: | Bcl-2 homologous antagonist/killer, human_BID_BH3_SAHB | | Authors: | Moldoveanu, T, Grace, C.R, Kriwacki, R.W, Green, D.R. | | Deposit date: | 2013-02-19 | | Release date: | 2013-04-17 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | BID-induced structural changes in BAK promote apoptosis.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2LX6
 
 | |
